using Plots
using DataInterpolations: LinearInterpolation
using StatsBase: Weights, sample
using Statistics: mean
using Random
using DisplayAs: PNG ## For faster rendering
Random.seed!(2024)Random.TaskLocalRNG()Stochastic chemical reaction: Gillespie Algorithm (direct and first reaction method) Adapted from: Chemical and Biomedical Engineering Calculations Using Python Ch.4-3
function ssa_alg(model, u0::AbstractVector, tend, p, stoich; tstart=zero(tend), method=:direct)
t = tstart ## Current time
ts = [t] ## Time points
u = copy(u0) ## Current state
us = copy(u') ## States over time
while t < tend
a = model(u, p, t) ## propensities
if method == :direct
dt = randexp() / sum(a) ## Time step for the direct method
du = sample(stoich, Weights(a)) ## Choose the stoichiometry for the next reaction
elseif method == :first
dts = randexp(length(a)) ./ a ## time scales of all reactions
i = argmin(dts) ## Choose the most recent reaction to occur
dt = dts[i]
du = stoich[i]
else
error("Method should be either :direct or :first")
end
u .+= du ## Update time
t += dt ## Update time
us = [us; u'] ## Append state
push!(ts, t) ## Append time point
end
return (t=ts, u=us)
endssa_alg (generic function with 1 method)Propensity model for this example reaction. Reaction of A <-> B with rate constants k1 & k2
Stoichiometry for each reaction
Initial conditions (Usually discrete values)
Simulation time
Solve the problem using both direct and first reaction method
0.685222 seconds (928.30 k allocations: 60.691 MiB, 19.67% gc time, 99.51% compilation time)
0.005784 seconds (16.35 k allocations: 14.937 MiB)(t = [0.0, 0.006874672879606227, 0.0070050915014185, 0.01329115182676454, 0.015335530116774018, 0.01942319769041779, 0.021346508038833215, 0.02345030020299408, 0.025497753122539372, 0.02734483697039726 … 9.937773197389458, 9.938360255993572, 9.964450740930666, 9.96564917153764, 9.975440668199402, 9.978594329657025, 9.982318425245289, 9.987213207313044, 9.995833233887383, 10.024199143896238], u = [200 0; 199 1; … ; 75 125; 74 126])Plot the solution from the direct method
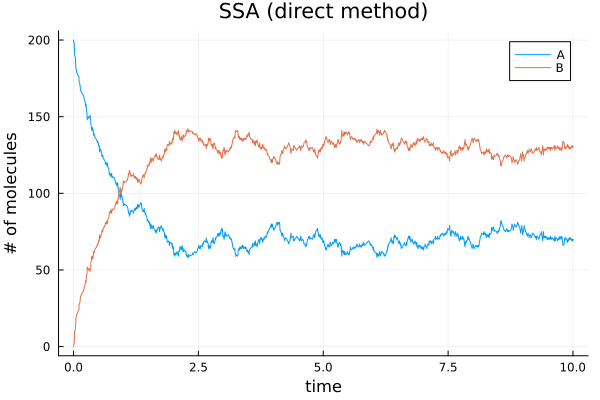
Plot the solution by the first reaction method
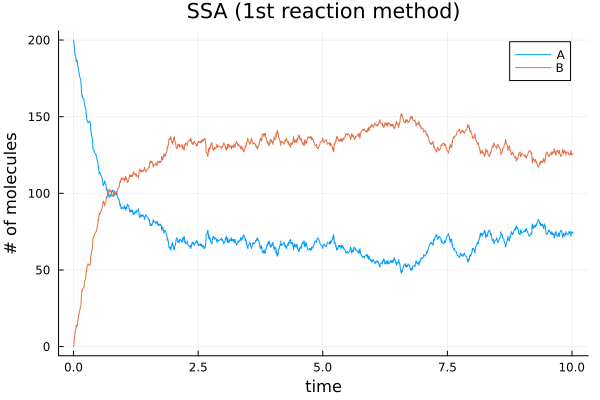
Running 50 simulations
0.181422 seconds (760.47 k allocations: 752.427 MiB, 24.61% gc time, 40.07% compilation time)Average values and interpolation
b_avg (generic function with 1 method)Plot the solution
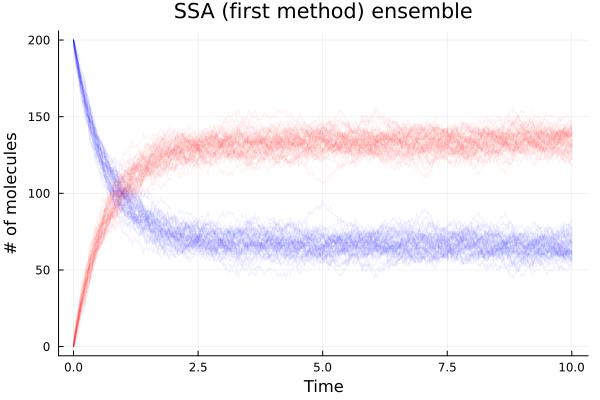
Plot averages
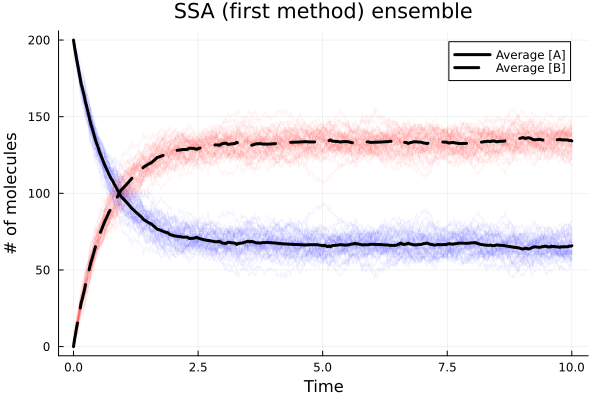
This notebook was generated using Literate.jl.