Fig 6.18#
Model of calcium-induced calcium release in hepatocytes
using Catalyst
using ModelingToolkit
using OrdinaryDiffEq
using Plots
Plots.default(linewidth=2)
rn = @reaction_network begin
@parameters I(t)
(k1 * I, km1), R <--> RI
(k2 * C, km2), RI <--> RIC
(k3 * C, km3), RIC <--> RICC
vr * (γ0 + γ1 * RIC) * (Cer - C), 0 --> C
hill(C, p1, p2, 4), C => 0
end
\[\begin{split} \begin{align*}
\mathrm{R} &\xrightleftharpoons[\mathtt{km1}]{\mathtt{k1} I\left( t \right)} \mathrm{\mathtt{RI}} \\
\mathrm{\mathtt{RI}} &\xrightleftharpoons[\mathtt{km2}]{C \mathtt{k2}} \mathrm{\mathtt{RIC}} \\
\mathrm{\mathtt{RIC}} &\xrightleftharpoons[\mathtt{km3}]{C \mathtt{k3}} \mathrm{\mathtt{RICC}} \\
\varnothing &\xrightleftharpoons[\frac{C^{4} \mathtt{p1}}{C^{4} + \mathtt{p2}^{4}}]{\left( - C + \mathtt{Cer} \right) \left( \mathtt{{\gamma}0} + \mathtt{RIC} \mathtt{{\gamma}1} \right) \mathtt{vr}} \mathrm{C}
\end{align*}
\end{split}\]
setdefaults!(rn, [
:γ0 => 0.1,
:γ1 => 20.5,
:p1 => 8.5,
:p2 => 0.065,
:k1 => 12,
:k2 => 15,
:k3 => 1.8,
:km1 => 8,
:km2 => 1.65,
:km3 => 0.21,
:Cer => 8.37,
:vr => 0.185,
:I => 0,
:C => 0,
:R => 1,
:RI => 0,
:RIC => 0,
:RICC => 0
])
osys = convert(ODESystem, rn; remove_conserved = true) |> structural_simplify
equations(osys)
\[\begin{split} \begin{align}
\frac{\mathrm{d} C\left( t \right)}{\mathrm{d}t} &= \frac{ - \left( C\left( t \right) \right)^{4} \mathtt{p1}}{\mathtt{p2}^{4} + \left( C\left( t \right) \right)^{4}} + \left( \mathtt{Cer} - C\left( t \right) \right) \mathtt{vr} \left( \mathtt{{\gamma}0} + \mathtt{RIC}\left( t \right) \mathtt{{\gamma}1} \right) \\
\frac{\mathrm{d} R\left( t \right)}{\mathrm{d}t} &= \mathtt{km1} \mathtt{RI}\left( t \right) - \mathtt{k1} R\left( t \right) I\left( t \right) \\
\frac{\mathrm{d} \mathtt{RI}\left( t \right)}{\mathrm{d}t} &= - \mathtt{km1} \mathtt{RI}\left( t \right) + \mathtt{km2} \mathtt{RIC}\left( t \right) + \mathtt{k1} R\left( t \right) I\left( t \right) - \mathtt{k2} \mathtt{RI}\left( t \right) C\left( t \right) \\
\frac{\mathrm{d} \mathtt{RIC}\left( t \right)}{\mathrm{d}t} &= - \mathtt{km2} \mathtt{RIC}\left( t \right) + \mathtt{km3} \left( - R\left( t \right) - \mathtt{RI}\left( t \right) - \mathtt{RIC}\left( t \right) + \Gamma_{1} \right) + \mathtt{k2} \mathtt{RI}\left( t \right) C\left( t \right) - \mathtt{k3} \mathtt{RIC}\left( t \right) C\left( t \right)
\end{align}
\end{split}\]
Fig 6.18 (A)#
@unpack I = osys
prob = ODEProblem(osys, [], (0., 25.), [I => 2.0])
sol = solve(prob)
@unpack C, RIC, RICC = osys
plot(sol, idxs=[C, RIC, RICC], title="Fig 6.18 (A)", xlabel="Time", ylabel="Abundance", legend=:topright)
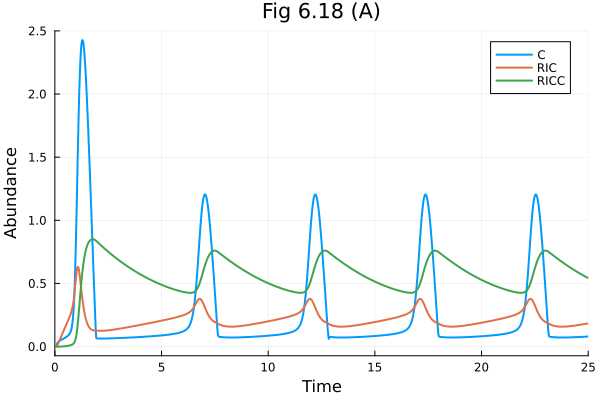
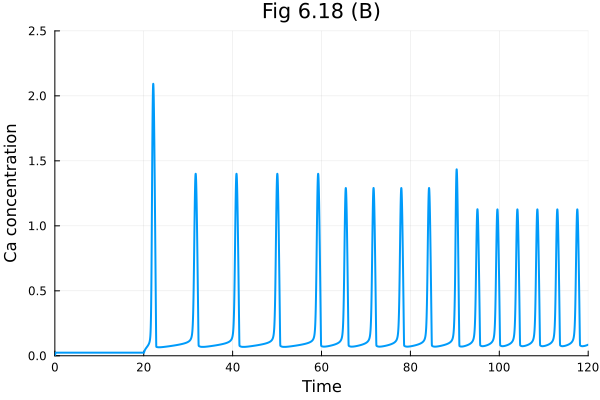
Fig 6.18 (B)#
discrete_events = [[20] => [I ~ 0.7], [60] => [I ~ 1.2], [90] => [I ~ 4.0]]
osys618 = convert(ODESystem, rn; discrete_events, remove_conserved = true) |> structural_simplify
tend = 120.
prob = ODEProblem(osys618, [], tend)
sol = solve(prob)
plot(sol, idxs=[osys.C], title="Fig 6.18 (B)", xlabel="Time", ylabel="Ca concentration", legend=false, ylim=(0, 2.5))
This notebook was generated using Literate.jl.

