using OrdinaryDiffEq
using Catalyst
using ModelingToolkit
using Plots
Plots.default(linewidth=2)Fig 2.09
Metabolic network simulation
Using Catalyst.jl to simulate a metabolic network.
rn = @reaction_network begin
k1, 0 --> A
k2, A --> B
k3, A + B --> C + D
k4, C --> 0
k5, D --> 0
end\[ \begin{align*} \varnothing &\xrightarrow{\mathtt{k1}} \mathrm{A} \\ \mathrm{A} &\xrightarrow{\mathtt{k2}} \mathrm{B} \\ \mathrm{A} + \mathrm{B} &\xrightarrow{\mathtt{k3}} \mathrm{C} + \mathrm{D} \\ \mathrm{C} &\xrightarrow{\mathtt{k4}} \varnothing \\ \mathrm{D} &\xrightarrow{\mathtt{k5}} \varnothing \end{align*} \]
Showing the differential equations in the reaction network
osys = convert(ODESystem, rn) |> complete
equations(osys)\[ \begin{align} \frac{\mathrm{d} A\left( t \right)}{\mathrm{d}t} &= \mathtt{k1} - \mathtt{k2} A\left( t \right) - \mathtt{k3} B\left( t \right) A\left( t \right) \\ \frac{\mathrm{d} B\left( t \right)}{\mathrm{d}t} &= \mathtt{k2} A\left( t \right) - \mathtt{k3} B\left( t \right) A\left( t \right) \\ \frac{\mathrm{d} C\left( t \right)}{\mathrm{d}t} &= - \mathtt{k4} C\left( t \right) + \mathtt{k3} B\left( t \right) A\left( t \right) \\ \frac{\mathrm{d} D\left( t \right)}{\mathrm{d}t} &= - \mathtt{k5} D\left( t \right) + \mathtt{k3} B\left( t \right) A\left( t \right) \end{align} \]
Solve the problem
ps = [:k1=>3., :k2=>2., :k3=>2.5, :k4=>3., :k5=>4.]
u0 = [:A=>0., :B=>0., :C=>0., :D=>0.]
tend = 10.
prob = ODEProblem(osys, u0, tend, ps)
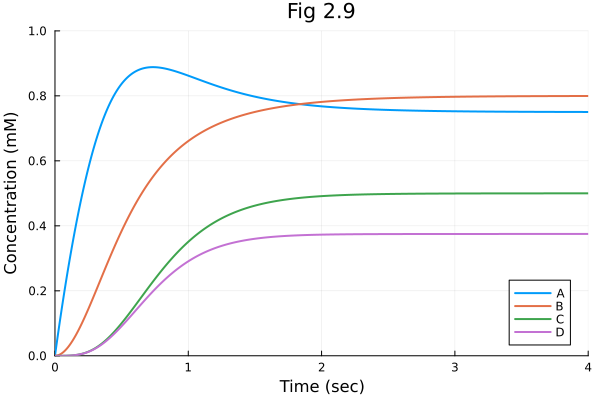
sol = solve(prob)retcode: Success
Interpolation: 3rd order Hermite
t: 31-element Vector{Float64}:
0.0
9.999999999999999e-5
0.0010999999999999998
0.011099999999999997
0.03427453254462915
0.06522753353833287
0.10153577459409753
0.14533869701822555
0.19788890020410435
0.2619461233746949
⋮
3.481141518838101
4.1462374627542244
4.984641398737613
5.978883857784344
6.9999177726334345
7.954887766924096
8.836082624890851
9.672663483036162
10.0
u: 31-element Vector{Vector{Float64}}:
[0.0, 0.0, 0.0, 0.0]
[0.00029997000199933764, 2.999799953754822e-8, 5.623912546644621e-16, 5.623800072204298e-16]
[0.003296372652316679, 3.6273312363558036e-6, 8.218056460901668e-12, 8.216249166278955e-12]
[0.0329330063952576, 0.0003668253522315969, 8.35676801381655e-8, 8.338285321893444e-8]
[0.09937122761544223, 0.0034375565665459304, 7.255794159499521e-6, 7.206618524863516e-6]
[0.1833655969433656, 0.012131166040593139, 8.934253880025109e-5, 8.820117939513387e-5]
[0.2751746900149442, 0.028399924487097824, 0.00048566621627305935, 0.00047610712983926046]
[0.3764608372444922, 0.055514839373642884, 0.0018497945329364433, 0.0017982532908780546]
[0.4844196905527862, 0.09656029732483219, 0.0056209605260374995, 0.0054101243611240964]
[0.5961814643349724, 0.1550531063006929, 0.0147148756470802, 0.013992428397168606]
⋮
[0.751015481451754, 0.7988628794584909, 0.4998345907732017, 0.374949157084143]
[0.7502915599254337, 0.7996724577481669, 0.499958490391288, 0.37498115400855275]
[0.7500631110964759, 0.7999305208686214, 0.4999887838283954, 0.37498954873657014]
[0.7500144275981989, 0.7999882359844388, 0.49999045642132917, 0.37497086078229874]
[0.7500125250580912, 0.7999980647430992, 0.49997804770113624, 0.3748883744178919]
[0.7500190130496543, 0.7999996560465515, 0.49996250817149734, 0.374746533274382]
[0.7500193831308702, 0.7999999319182715, 0.49996133873867094, 0.3746770632490242]
[0.7500144341539133, 0.7999999855119406, 0.499971154272474, 0.3747145705226947]
[0.7500039199737626, 0.7999999921573426, 0.4999921725366447, 0.37492503636481844]Visual
plot(sol, legend=:bottomright, title="Fig 2.9",
xlims=(0., 4.), ylims=(0., 1.),
xlabel="Time (sec)", ylabel="Concentration (mM)"
)
This notebook was generated using Literate.jl.
Back to top