using OrdinaryDiffEq
using Plots
Plots.default(linewidth=2)Solving differential equations in Julia
Standard procedures
- Define a model function representing the right-hand-side (RHS) of the system.
- Out-of-place form:
f(u, p, t)whereuis the state variable(s),pis the parameter(s), andtis the independent variable (usually time). The output is the right hand side (RHS) of the differential equation system. - In-place form:
f!(du, u, p, t), where the output is saved todu. The rest is the same as the out of place form. The in-place form has potential performance benefits since it allocates less than the out-of-place (f(u, p, t)) counterpart. - Using ModelingToolkit.jl : define equations and build an ODE system.
- Out-of-place form:
- Initial conditions (
u0) for the state variable(s). - (Optional) define parameter(s)
p. - Define a problem (e.g.
ODEProblem) using the modeling function (f), initial conditions (u0), simulation time span (tspan == (tstart, tend)), and parameter(s)p. - Solve the problem by calling
solve(prob).
Solve ODEs using OrdinaryDiffEq.jl
Documentation: https://docs.sciml.ai/DiffEqDocs/stable/
Single variable: Exponential decay model
The concentration of a decaying nuclear isotope could be described as an exponential decay:
\[ \frac{d}{dt}C(t) = - \lambda C(t) \]
State variable - \(C(t)\): The concentration of a decaying nuclear isotope.
Parameter - \(\lambda\): The rate constant of decay. The half-life \(t_{\frac{1}{2}} = \frac{ln2}{\lambda}\)
The model function is the 3-argument out-of-place form, f(u, p, t).
decay(u, p, t) = p * u
p = -1.0 ## Rate of exponential decay
u0 = 1.0 ## Initial condition
tspan = (0.0, 2.0) ## Start time and end time
prob = ODEProblem(decay, u0, tspan, p)
sol = solve(prob)retcode: Success
Interpolation: 3rd order Hermite
t: 8-element Vector{Float64}:
0.0
0.10001999200479662
0.34208427066999536
0.6553980136343391
1.0312652525315806
1.4709405856363595
1.9659576669700232
2.0
u: 8-element Vector{Float64}:
1.0
0.9048193287657775
0.7102883621328674
0.5192354400036404
0.3565557657699655
0.22970979078638265
0.1400224727245278
0.13533600284008826Solution at time t=1.0 (with interpolation)
sol(1.0)0.3678796381978344Time points
sol.t8-element Vector{Float64}:
0.0
0.10001999200479662
0.34208427066999536
0.6553980136343391
1.0312652525315806
1.4709405856363595
1.9659576669700232
2.0Solutions at corresponding time points
sol.u8-element Vector{Float64}:
1.0
0.9048193287657775
0.7102883621328674
0.5192354400036404
0.3565557657699655
0.22970979078638265
0.1400224727245278
0.13533600284008826Visualize the solution
plot(sol)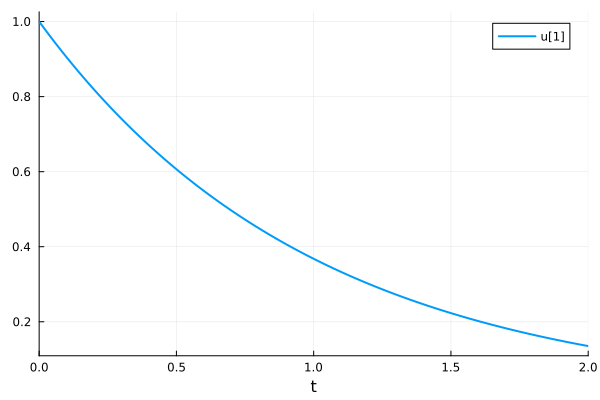
Three variables: The SIR model
The SIR model describes the spreading of an contagious disease can be described by the SIR model:
\[ \begin{align} \frac{d}{dt}S(t) &= - \beta S(t)I(t) \\ \frac{d}{dt}I(t) &= \beta S(t)I(t) - \gamma I(t) \\ \frac{d}{dt}R(t) &= \gamma I(t) \end{align} \]
State variables
- \(S(t)\) : the fraction of susceptible people
- \(I(t)\) : the fraction of infectious people
- \(R(t)\) : the fraction of recovered (or removed) people
Parameters
- \(\beta\) : the rate of infection when susceptible and infectious people meet
- \(\gamma\) : the rate of recovery of infectious people
using OrdinaryDiffEq
using Plots
Plots.default(linewidth=2)SIR model (in-place form can save array allocations and thus faster)
function sir!(du, u, p, t)
s, i, r = u
β, γ = p
v1 = β * s * i
v2 = γ * i
du[1] = -v1
du[2] = v1 - v2
du[3] = v2
return nothing
endsir! (generic function with 1 method)p = (β=1.0, γ=0.3)
u0 = [0.99, 0.01, 0.00]
tspan = (0.0, 20.0)
prob = ODEProblem(sir!, u0, tspan, p)
sol = solve(prob)retcode: Success
Interpolation: 3rd order Hermite
t: 17-element Vector{Float64}:
0.0
0.08921318693905476
0.3702862715172094
0.7984257132319627
1.3237271485666187
1.991841832691831
2.7923706947355837
3.754781614278828
4.901904318934307
6.260476636498209
7.7648912410433075
9.39040980993922
11.483861023017885
13.372369854616487
15.961357172044833
18.681426667664056
20.0
u: 17-element Vector{Vector{Float64}}:
[0.99, 0.01, 0.0]
[0.9890894703413342, 0.010634484617786016, 0.00027604504087978485]
[0.9858331594901347, 0.012901496825852227, 0.0012653436840130792]
[0.9795270529591532, 0.017282420996456258, 0.003190526044390598]
[0.9689082167415561, 0.024631267034445452, 0.006460516223998509]
[0.9490552312363142, 0.03827338797605376, 0.012671380787632129]
[0.911862947533394, 0.06347250098224955, 0.024664551484356523]
[0.8398871089274514, 0.11078176031568526, 0.04933113075686341]
[0.707584206802473, 0.19166147882272797, 0.10075431437479916]
[0.5081460287219878, 0.2917741934147055, 0.2000797778633069]
[0.31213222024414106, 0.3415879120018043, 0.34627986775405484]
[0.1821568309636563, 0.309998313415639, 0.5078448556207048]
[0.10427205468919257, 0.2206111401113331, 0.6751168051994745]
[0.07386737407725885, 0.14760143051851196, 0.7785311954042293]
[0.05545028910907742, 0.07997076922865369, 0.864578941662269]
[0.04733499069589219, 0.040605653213833574, 0.9120593560902743]
[0.04522885458929347, 0.02905741611081477, 0.9257137292998918]Visualize the solution
plot(sol, label=["S" "I" "R"], legend=:right)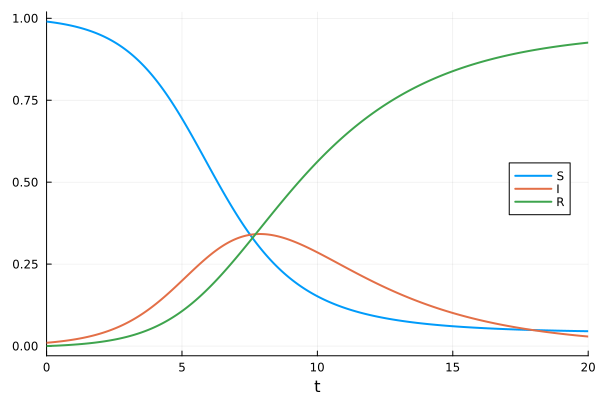
Using ModelingToolkit.jl (recommended)
ModelingToolkit.jl is a high-level package for symbolic-numeric modeling and simulation in the Julia ecosystem.
using ModelingToolkit
using OrdinaryDiffEq
using Plots
Plots.default(linewidth=2)Exponential decay model
@independent_variables t ## Time
@parameters λ ## Decaying rate constant
@variables C(t) ## Time and concentration
D = Differential(t) ## Differential operatorDifferential(t)Define an ODE with equations
eqs = [D(C) ~ -λ * C]
@mtkbuild decaySys = ODESystem(eqs, t)\[ \begin{align} \frac{\mathrm{d} C\left( t \right)}{\mathrm{d}t} &= - C\left( t \right) \lambda \end{align} \]
u0 = [C => 1.0]
p = [λ => 1.0]
tspan = (0.0, 2.0)
prob = ODEProblem(decaySys, u0, tspan, p)
sol = solve(prob)
plot(sol)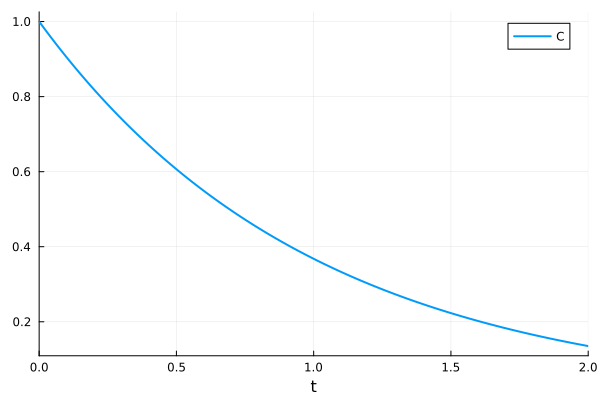
SIR model
using OrdinaryDiffEq
using ModelingToolkit
using Plots
Plots.default(linewidth=2)
@independent_variables t
@parameters β γ
@variables s(t) i(t) r(t)
D = Differential(t)
eqs = [
D(s) ~ -β * s * i,
D(i) ~ β * s * i - γ * i,
D(r) ~ γ * i
]
@mtkbuild sirSys = ODESystem(eqs, t)\[ \begin{align} \frac{\mathrm{d} i\left( t \right)}{\mathrm{d}t} &= - i\left( t \right) \gamma + i\left( t \right) s\left( t \right) \beta \\ \frac{\mathrm{d} r\left( t \right)}{\mathrm{d}t} &= i\left( t \right) \gamma \\ \frac{\mathrm{d} s\left( t \right)}{\mathrm{d}t} &= - i\left( t \right) s\left( t \right) \beta \end{align} \]
p = [β => 1.0, γ => 0.3]
u0 = [s => 0.99, i => 0.01, r => 0.00]
tspan = (0.0, 20.0)
prob = ODEProblem(sirSys, u0, tspan, p)
sol = solve(prob)
plot(sol)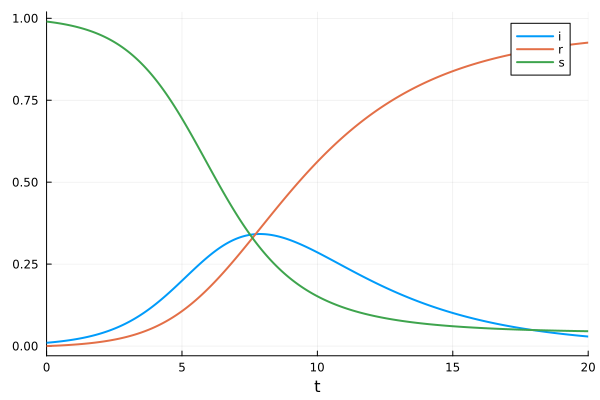
Lorenz system
The Lorenz system is a system of ordinary differential equations having chaotic solutions for certain parameter values and initial conditions. (Wikipedia)
\[ \begin{align} \frac{dx}{dt} &= \sigma(y-x) \\ \frac{dy}{dt} &= x(\rho - z) -y \\ \frac{dz}{dt} &= xy - \beta z \end{align} \]
using OrdinaryDiffEq
using ModelingToolkit
using Plots
Plots.default(linewidth=2)Building the model is wrapped in a function
function build_lorentz(; name)
@parameters begin
σ = 10.0
ρ = 28.0
β = 8 / 3
end
@independent_variables t
@variables begin
x(t) = 1.0 ## Independent variable (time)
y(t) = 0.0 ## Independent variable (time)
z(t) = 0.0 ## Independent variable (time)
end
D = Differential(t)
eqs = [
D(x) ~ σ * (y - x),
D(y) ~ x * (ρ - z) - y,
D(z) ~ x * y - β * z
]
sys = ODESystem(eqs, t; name)
return sys
endbuild_lorentz (generic function with 1 method)tspan = (0.0, 100.0)
@mtkbuild sys = build_lorentz()
prob = ODEProblem(sys, [], tspan, [])
sol = solve(prob)retcode: Success
Interpolation: 3rd order Hermite
t: 1325-element Vector{Float64}:
0.0
3.5678604836301404e-5
0.0003924646531993154
0.003262407732952926
0.009058075632950364
0.01695646917889552
0.027689958632766257
0.04185634991045167
0.06024041181372633
0.08368541086439041
⋮
99.49018875318062
99.54862803586417
99.6145902246956
99.6844772275333
99.76689729728005
99.85118113701569
99.9277902563632
99.99633045999587
100.0
u: 1325-element Vector{Vector{Float64}}:
[1.0, 0.0, 0.0]
[0.9996434557625105, 0.0009988049817849058, 1.781434788799208e-8]
[0.9961045497425811, 0.010965399721242457, 2.146955365838907e-6]
[0.9693591636090403, 0.08977060609621872, 0.0001438018323641714]
[0.9242043615200224, 0.24228912476918024, 0.001046162329727883]
[0.8800455859164964, 0.4387364554855289, 0.003424259429099474]
[0.8483309846305274, 0.6915629338584358, 0.008487624633044172]
[0.8495036661237968, 1.0145426239346456, 0.018212089206688865]
[0.9139069582771231, 1.4425599843977082, 0.036693822160421524]
[1.0888638105810242, 2.0523265477022825, 0.07402573342120332]
⋮
[-0.9940591016433658, -2.8817300441171474, 21.71504831029168]
[-1.928880273667646, -3.388601591720348, 18.833364484674814]
[-2.9751110649395445, -4.815097635912501, 16.400650729866644]
[-4.625266725547489, -7.632665256875718, 15.107297022943598]
[-7.9439838455455565, -13.030768900739405, 16.94732621617739]
[-12.5991630525995, -17.606167341053442, 26.084946638181943]
[-14.18564328360291, -12.357314081660197, 36.407114774499064]
[-10.767406782468159, -3.6671228558902, 36.59399970621433]
[-10.505291991542054, -3.323107353088182, 36.373367127901254]x-y-z time-series
plot(sol)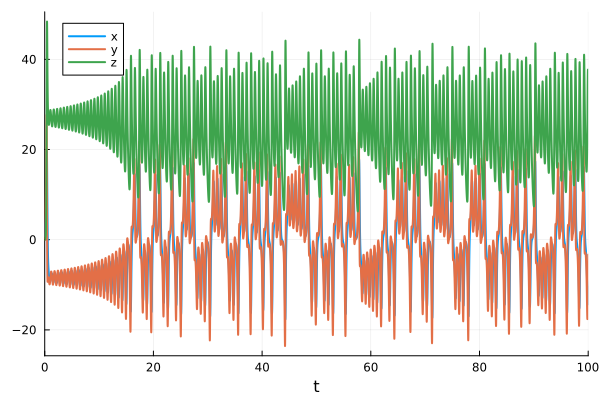
y-t plot
plot(sol, idxs=[sys.y])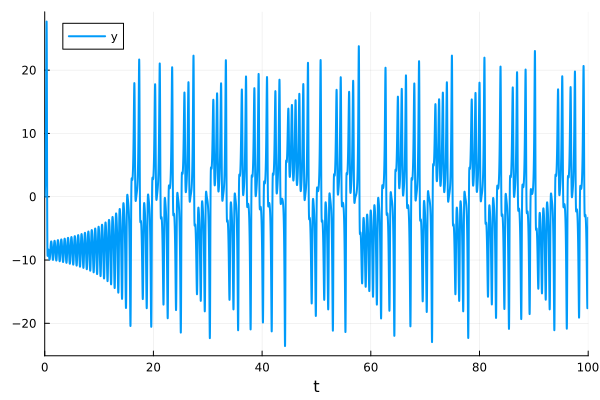
idxs=(sys.x, sys.y, sys.z) makes a phase plot.
plot(sol, idxs=(sys.x, sys.y, sys.z), label=false, size=(600, 600))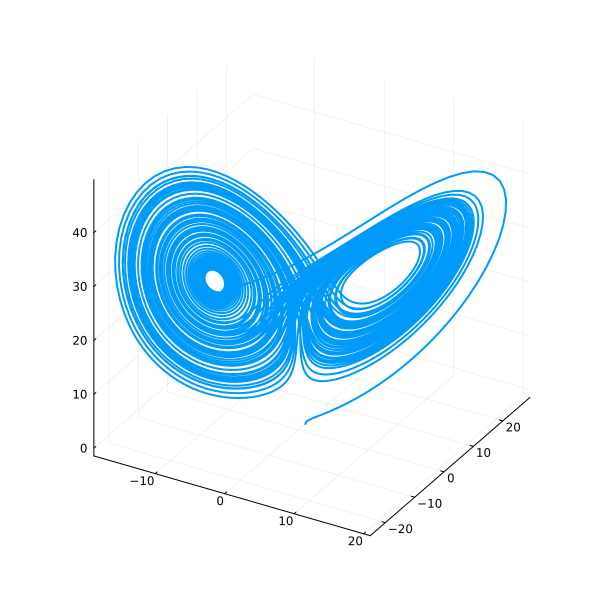
Saving simulation results
using DataFrames
using CSV
df = DataFrame(sol)
CSV.write("lorenz.csv", df)
rm("lorenz.csv")Catalyst.jl
Catalyst.jl is a domain-specific language (DSL) package to simulate chemical reaction networks.
using OrdinaryDiffEq
using Catalyst
using Plots
Plots.default(linewidth=2)Exponential decay model
decay_rn = @reaction_network begin
λ, C --> 0
end\[ \begin{align*} \mathrm{C} &\xrightarrow{\lambda} \varnothing \end{align*} \]
p = [:λ => 1.0]
u0 = [:C => 1.0]
tspan = (0.0, 2.0)
prob = ODEProblem(decay_rn, u0, tspan, p)
sol = solve(prob)retcode: Success
Interpolation: 3rd order Hermite
t: 8-element Vector{Float64}:
0.0
0.10001999200479662
0.34208427066999536
0.6553980136343391
1.0312652525315806
1.4709405856363595
1.9659576669700232
2.0
u: 8-element Vector{Vector{Float64}}:
[1.0]
[0.9048193287657775]
[0.7102883621328674]
[0.5192354400036404]
[0.3565557657699655]
[0.22970979078638265]
[0.1400224727245278]
[0.13533600284008826]plot(sol, title="Exponential Decay")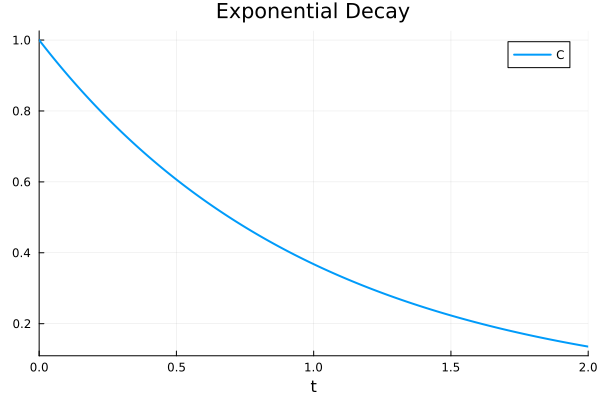
SIR model
sir_rn = @reaction_network begin
β, S + I --> 2I
γ, I --> R
end\[ \begin{align*} \mathrm{S} + \mathrm{I} &\xrightarrow{\beta} 2 \mathrm{I} \\ \mathrm{I} &\xrightarrow{\gamma} \mathrm{R} \end{align*} \]
Extract the symbols for later use
@unpack β, γ, S, I, R = sir_rn
p = [β => 1.0, γ => 0.3]
u0 = [S => 0.99, I => 0.01, R => 0.00]
tspan = (0.0, 20.0)
prob = ODEProblem(sir_rn, u0, tspan, p)
sol = solve(prob)retcode: Success
Interpolation: 3rd order Hermite
t: 17-element Vector{Float64}:
0.0
0.08921318693905476
0.3702862715172094
0.7984257132319627
1.3237271485666187
1.991841832691831
2.7923706947355837
3.754781614278828
4.901904318934307
6.260476636498209
7.7648912410433075
9.39040980993922
11.483861023017885
13.372369854616487
15.961357172044833
18.681426667664056
20.0
u: 17-element Vector{Vector{Float64}}:
[0.99, 0.01, 0.0]
[0.9890894703413342, 0.010634484617786016, 0.00027604504087978485]
[0.9858331594901347, 0.012901496825852227, 0.0012653436840130792]
[0.9795270529591532, 0.017282420996456258, 0.003190526044390598]
[0.9689082167415561, 0.024631267034445452, 0.006460516223998509]
[0.9490552312363142, 0.03827338797605376, 0.012671380787632129]
[0.911862947533394, 0.06347250098224955, 0.024664551484356523]
[0.8398871089274514, 0.11078176031568526, 0.04933113075686341]
[0.707584206802473, 0.19166147882272797, 0.10075431437479916]
[0.5081460287219878, 0.2917741934147055, 0.2000797778633069]
[0.31213222024414106, 0.3415879120018043, 0.34627986775405484]
[0.1821568309636563, 0.309998313415639, 0.5078448556207048]
[0.10427205468919257, 0.2206111401113331, 0.6751168051994745]
[0.07386737407725885, 0.14760143051851196, 0.7785311954042293]
[0.05545028910907742, 0.07997076922865369, 0.864578941662269]
[0.04733499069589219, 0.040605653213833574, 0.9120593560902743]
[0.04522885458929347, 0.02905741611081477, 0.9257137292998918]plot(sol, legend=:right, title="SIR Model")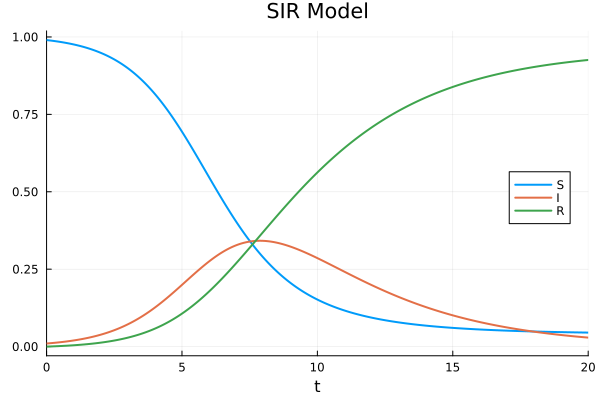
This notebook was generated using Literate.jl.