using ModelingToolkit
using Catalyst
using OrdinaryDiffEq
using DiffEqCallbacks
using SteadyStateDiffEq
using Plots
Plots.default(linewidth=2)Fig 6.5
Model of G-protein signalling pathway
rn = @reaction_network begin
@parameters L(t)
(kRL * L, kRLm), R <--> RL
kGa, G + RL --> Ga + Gbg + RL
kGd0, Ga --> Gd
kG1, Gd + Gbg --> G
end\[ \begin{align*} \mathrm{R} &\xrightleftharpoons[\mathtt{kRLm}]{\mathtt{kRL} L\left( t \right)} \mathrm{\mathtt{RL}} \\ \mathrm{G} + \mathrm{\mathtt{RL}} &\xrightarrow{\mathtt{kGa}} \mathrm{\mathtt{Ga}} + \mathrm{\mathtt{Gbg}} + \mathrm{\mathtt{RL}} \\ \mathrm{\mathtt{Ga}} &\xrightarrow{\mathtt{kGd0}} \mathrm{\mathtt{Gd}} \\ \mathrm{\mathtt{Gd}} + \mathrm{\mathtt{Gbg}} &\xrightarrow{\mathtt{kG1}} \mathrm{G} \end{align*} \]
setdefaults!(rn, [
:kRL => 2e6,
:kRLm => 0.01,
:kGa => 1e-5,
:kGd0 => 0.11,
:kG1 => 1,
:R => 4e3,
:RL => 0.,
:G => 1e4,
:Ga => 0.,
:Gd => 0.,
:Gbg => 0.,
:L => 0.
])
@unpack L = rn
discrete_events = [[200] => [L~1e-9], [800] => [L~0.0]]
osys = convert(ODESystem, rn; discrete_events, remove_conserved = true) |> complete\[ \begin{align} \frac{\mathrm{d} R\left( t \right)}{\mathrm{d}t} &= \mathtt{kRLm} \left( - R\left( t \right) + \Gamma_{1} \right) - \mathtt{kRL} R\left( t \right) L\left( t \right) \\ \frac{\mathrm{d} G\left( t \right)}{\mathrm{d}t} &= \mathtt{kG1} \left( - G\left( t \right) + \Gamma_{2} \right) \left( - G\left( t \right) - \mathtt{Ga}\left( t \right) + \Gamma_{3} \right) - \mathtt{kGa} \left( - R\left( t \right) + \Gamma_{1} \right) G\left( t \right) \\ \frac{\mathrm{d} \mathtt{Ga}\left( t \right)}{\mathrm{d}t} &= - \mathtt{kGd0} \mathtt{Ga}\left( t \right) + \mathtt{kGa} \left( - R\left( t \right) + \Gamma_{1} \right) G\left( t \right) \end{align} \]
Fig 6.5 A
tend = 1200.0
prob = ODEProblem(osys, [], tend)
sol = solve(prob, Rodas5P(), abstol=1e-8, reltol=1e-8)
@unpack RL, Ga = osys
plot(sol, idxs=[RL, Ga], title="Fig 6.05 (A)", xlabel="Time", ylabel="Abundance")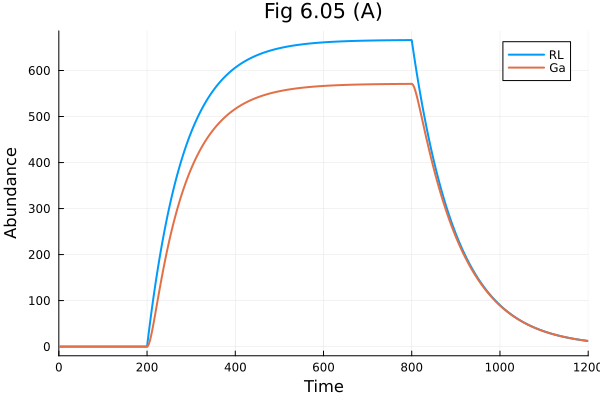
Fig 6.5 B
lrange = range(0, 20 * 1e-9, 101)
prob_func = (prob, i, repeat) -> remake(prob, p=[L => lrange[i]])
prob = SteadyStateProblem(osys, [], [])
trajectories = length(lrange)
alg = DynamicSS(Rodas5())
eprob = EnsembleProblem(prob; prob_func)
sim = solve(eprob, alg; trajectories, abstol=1e-10, reltol=1e-10)
ga = map(s->s[Ga], sim)
rl = map(s->s[RL], sim)
plot(lrange .* 1e9, [ga rl], label=["Ga" "RL"], title="Fig. 6.5 (B)",
xlabel="Ligand (nM)", ylabel="Steady-state abundance", xlims=(0, 20), ylims=(0, 3500))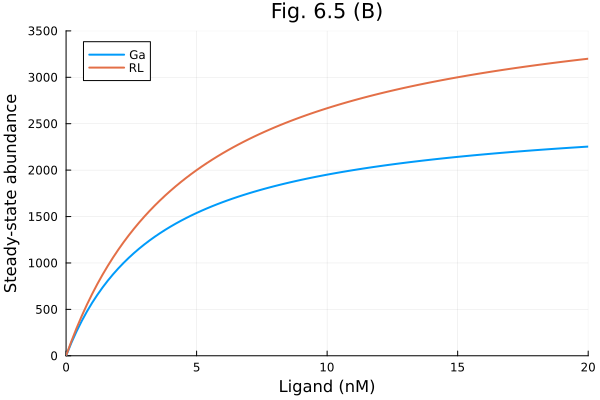
This notebook was generated using Literate.jl.