Calcium overload#
Steady-state solutions across a range of glucose levels.
using OrdinaryDiffEq
using SteadyStateDiffEq
using ModelingToolkit
using MitochondrialDynamics
using MitochondrialDynamics: μM
import PythonPlot as plt
plt.matplotlib.rcParams["font.size"] = 14
14
Default model
@named sys = make_model()
prob = SteadyStateProblem(sys, [])
alg = DynamicSS(Rodas5())
sol = solve(prob, alg)
retcode: Success
u: 9-element Vector{Float64}:
0.05710335793593597
0.2424229695492959
0.5365859320570429
0.006798790390977675
0.00247206877315645
0.08901096138926226
0.00019466055321005286
0.0008000956847624295
0.050077868373645945
High calcium model
@unpack RestingCa, ActivatedCa = sys
prob_ca5 = SteadyStateProblem(sys, [], [RestingCa=>0.45μM, ActivatedCa=>1.25μM])
prob_ca10 = SteadyStateProblem(sys, [], [RestingCa=>0.9μM, ActivatedCa=>2.5μM])
┌ Warning: `SciMLBase.SteadyStateProblem(sys, u0, p; kw...)` is deprecated. Use `SciMLBase.SteadyStateProblem(sys, merge(if isempty(u0)
│ Dict()
│ else
│ Dict(unknowns(sys) .=> u0)
│ end, Dict(p)))`
│ instead.
└ @ ModelingToolkit ~/.julia/packages/ModelingToolkit/b28X4/src/deprecations.jl:127
┌ Warning: `SciMLBase.SteadyStateProblem(sys, u0, p; kw...)` is deprecated. Use `SciMLBase.SteadyStateProblem(sys, merge(if isempty(u0)
│ Dict()
│ else
│ Dict(unknowns(sys) .=> u0)
│ end, Dict(p)))`
│ instead.
└ @ ModelingToolkit ~/.julia/packages/ModelingToolkit/b28X4/src/deprecations.jl:127
SteadyStateProblem with uType Vector{Float64}. In-place: true
u0: 9-element Vector{Float64}:
0.06
0.24
0.9
0.0087
0.0029
0.092
0.0002
0.001
0.057
Simulating on a range of glucose
@unpack Glc = sys
\[\begin{split} \begin{align}
\frac{\mathrm{d} \mathtt{x3}\left( t \right)}{\mathrm{d}t} &= \mathtt{tipside}\left( t \right) \\
\frac{\mathrm{d} \mathtt{x2}\left( t \right)}{\mathrm{d}t} &= - \mathtt{tipside}\left( t \right) + \mathtt{tiptip}\left( t \right) \\
\frac{\mathrm{d} \mathtt{AEC}\left( t \right)}{\mathrm{d}t} &= \frac{\mathtt{J\_ANT}\left( t \right) + 2 \mathtt{J\_GPD}\left( t \right) - \mathtt{ATPstiochGK} \mathtt{J\_GK}\left( t \right) + \left( - \mathtt{kATP} - \mathtt{kATPCa} \mathtt{Ca\_c}\left( t \right) \right) \mathtt{ATP\_c}\left( t \right)}{\mathtt{V\_I} \mathtt{{\Sigma}Ac}} \\
\frac{\mathrm{d} \mathtt{Pyr}\left( t \right)}{\mathrm{d}t} &= \frac{ - \mathtt{J\_LDH}\left( t \right) - \mathtt{J\_PDH}\left( t \right) + \mathtt{J\_GPD}\left( t \right)}{\mathtt{V\_I} + \mathtt{V\_MTX}} - \mathtt{kPyr} \mathtt{Pyr}\left( t \right) \\
\frac{\mathrm{d} \mathtt{G3P}\left( t \right)}{\mathrm{d}t} &= \frac{ - \mathtt{J\_GPD}\left( t \right) + 2 \mathtt{J\_GK}\left( t \right)}{\mathtt{V\_I}} - \mathtt{kG3P} \mathtt{G3P}\left( t \right) \\
\frac{\mathrm{d} \mathtt{\Delta{\Psi}m}\left( t \right)}{\mathrm{d}t} &= \frac{ - 2 \mathtt{J\_MCU}\left( t \right) + \mathtt{J\_HR}\left( t \right) - \mathtt{J\_HL}\left( t \right) - \mathtt{J\_HF}\left( t \right) - \mathtt{J\_ANT}\left( t \right)}{\mathtt{C\_MIT}} \\
\frac{\mathrm{d} \mathtt{Ca\_m}\left( t \right)}{\mathrm{d}t} &= \frac{\mathtt{F\_M} \left( \mathtt{J\_MCU}\left( t \right) - \mathtt{J\_NCLX}\left( t \right) \right)}{\mathtt{V\_MTX}} \\
\frac{\mathrm{d} \mathtt{NADH\_c}\left( t \right)}{\mathrm{d}t} &= \frac{ - \mathtt{J\_LDH}\left( t \right) - \mathtt{J\_NADHT}\left( t \right) + \mathtt{J\_GPD}\left( t \right)}{\mathtt{V\_I}} - \mathtt{kNADHc} \mathtt{NADH\_c}\left( t \right) \\
\frac{\mathrm{d} \mathtt{NADH\_m}\left( t \right)}{\mathrm{d}t} &= \frac{\mathtt{J\_DH}\left( t \right) + \mathtt{J\_NADHT}\left( t \right) - \mathtt{J\_O2}\left( t \right)}{\mathtt{V\_MTX}} - \mathtt{kNADHm} \mathtt{NADH\_m}\left( t \right)
\end{align}
\end{split}\]
Test on a range of glucose
glc = 3.5:0.5:30.0
prob_func = (prob, i, repeat) -> begin
remake(prob, p=[Glc => glc[i]])
end
trajectories=length(glc)
sim = solve(EnsembleProblem(prob; prob_func, safetycopy=false), alg; trajectories)
sim_ca5 = solve(EnsembleProblem(prob_ca5; prob_func, safetycopy=false), alg; trajectories)
sim_ca10 = solve(EnsembleProblem(prob_ca10; prob_func, safetycopy=false), alg; trajectories);
Steady states for a range of glucose#
function plot_steady_state(glc, sols, sys; figsize=(10, 10), title="")
@unpack G3P, Pyr, Ca_c, Ca_m, NADH_c, NADH_m, NAD_c, NAD_m, ATP_c, ADP_c, AMP_c, ΔΨm, x1, x2, x3, degavg = sys
glc5 = glc ./ 5
g3p = extract(sols, G3P * 1000)
pyr = extract(sols, Pyr * 1000)
ca_c = extract(sols, Ca_c * 1000)
ca_m = extract(sols, Ca_m * 1000)
nad_ratio_c = extract(sols, NADH_c/NAD_c)
nad_ratio_m = extract(sols, NADH_m/NAD_m)
atp_c = extract(sols, ATP_c * 1000)
adp_c = extract(sols, ADP_c * 1000)
amp_c = extract(sols, AMP_c * 1000)
td = extract(sols, ATP_c / ADP_c)
dpsi = extract(sols, ΔΨm * 1000)
x1 = extract(sols, x1)
x2 = extract(sols, x2)
x3 = extract(sols, x3)
deg = extract(sols, degavg)
numrows = 3
numcols = 3
fig, ax = plt.subplots(numrows, numcols; figsize)
ax[0, 0].plot(glc5, g3p)
ax[0, 0].set(ylabel="G3P (μM)")
ax[0, 0].set_title("a", loc="left")
ax[0, 1].plot(glc5, pyr)
ax[0, 1].set(ylabel="Pyruvate (μM)")
ax[0, 1].set_title("b", loc="left")
ax[0, 2].plot(glc5, ca_c, label="cyto")
ax[0, 2].plot(glc5, ca_m, label="mito")
ax[0, 2].legend()
ax[0, 2].set(ylabel="Calcium (μM)")
ax[0, 2].set_title("c", loc="left")
ax[1, 0].plot(glc5, nad_ratio_c, label="cyto")
ax[1, 0].plot(glc5, nad_ratio_m, label="mito")
ax[1, 0].legend()
ax[1, 0].set(ylabel="NADH:NAD (ratio)")
ax[1, 0].set_title("d", loc="left")
ax[1, 1].plot(glc5, atp_c, label="ATP")
ax[1, 1].plot(glc5, adp_c, label="ADP")
ax[1, 1].plot(glc5, amp_c, label="AMP")
ax[1, 1].legend()
ax[1, 1].set(ylabel="Adenylates (μM)")
ax[1, 1].set_title("e", loc="left")
ax[1, 2].plot(glc5, td)
ax[1, 2].set(ylabel="ATP:ADP (ratio)")
ax[1, 2].set_title("f", loc="left")
ax[2, 0].plot(glc5, dpsi, label="cyto")
ax[2, 0].set(ylabel="ΔΨ (mV)", xlabel="Glucose (X)")
ax[2, 0].set_title("g", loc="left")
ax[2, 1].plot(glc5, x1, label="X1")
ax[2, 1].plot(glc5, x2, label="X2")
ax[2, 1].plot(glc5, x3, label="X3")
ax[2, 1].set(ylabel="Mitochondrial nodes (a.u.)", xlabel="Glucose (X)")
ax[2, 1].set_title("h", loc="left")
ax[2, 2].plot(glc5, deg)
ax[2, 2].set(ylabel="Avg. Node Degree (a.u.)", xlabel="Glucose (X)")
ax[2, 2].set_title("i", loc="left")
for i in 0:numrows-1, j in 0:numcols-1
ax[i, j].set_xticks(1:6)
ax[i, j].grid()
end
fig.suptitle(title)
fig.tight_layout()
return fig
end
plot_steady_state (generic function with 1 method)
Default model
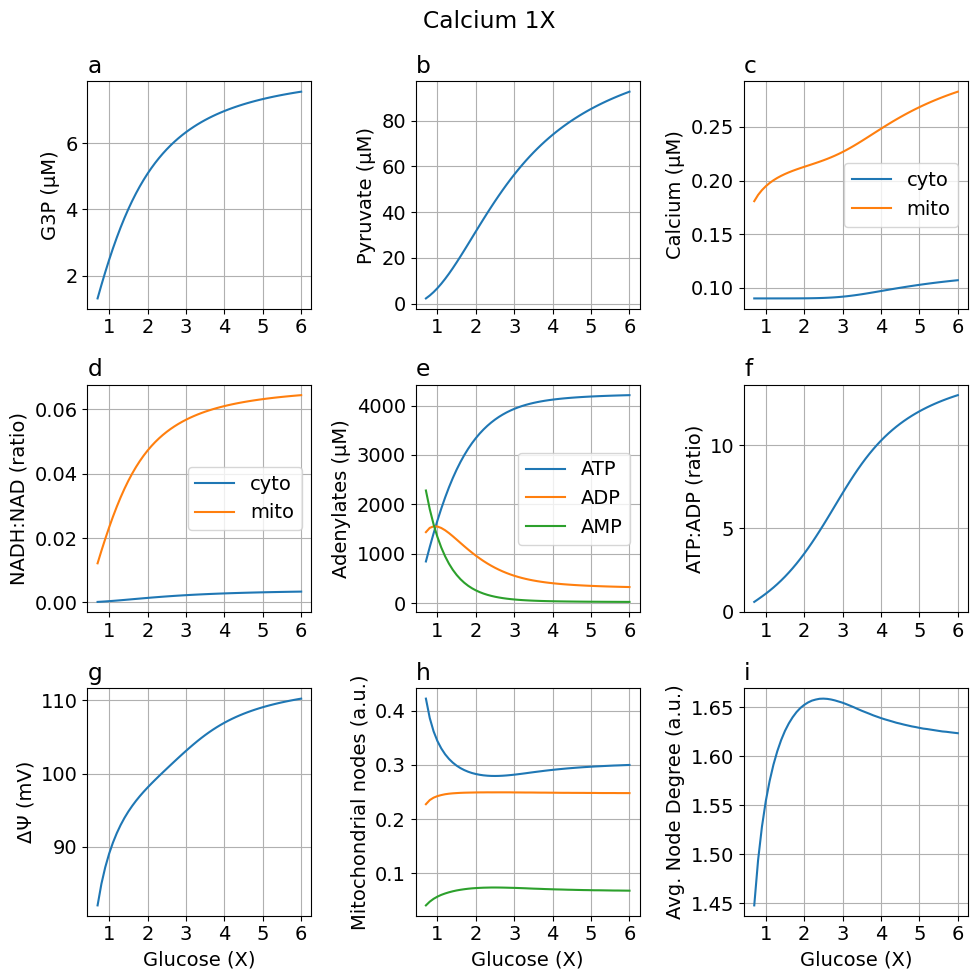
fig_glc_default = plot_steady_state(glc, sim, sys, title="Calcium 1X")
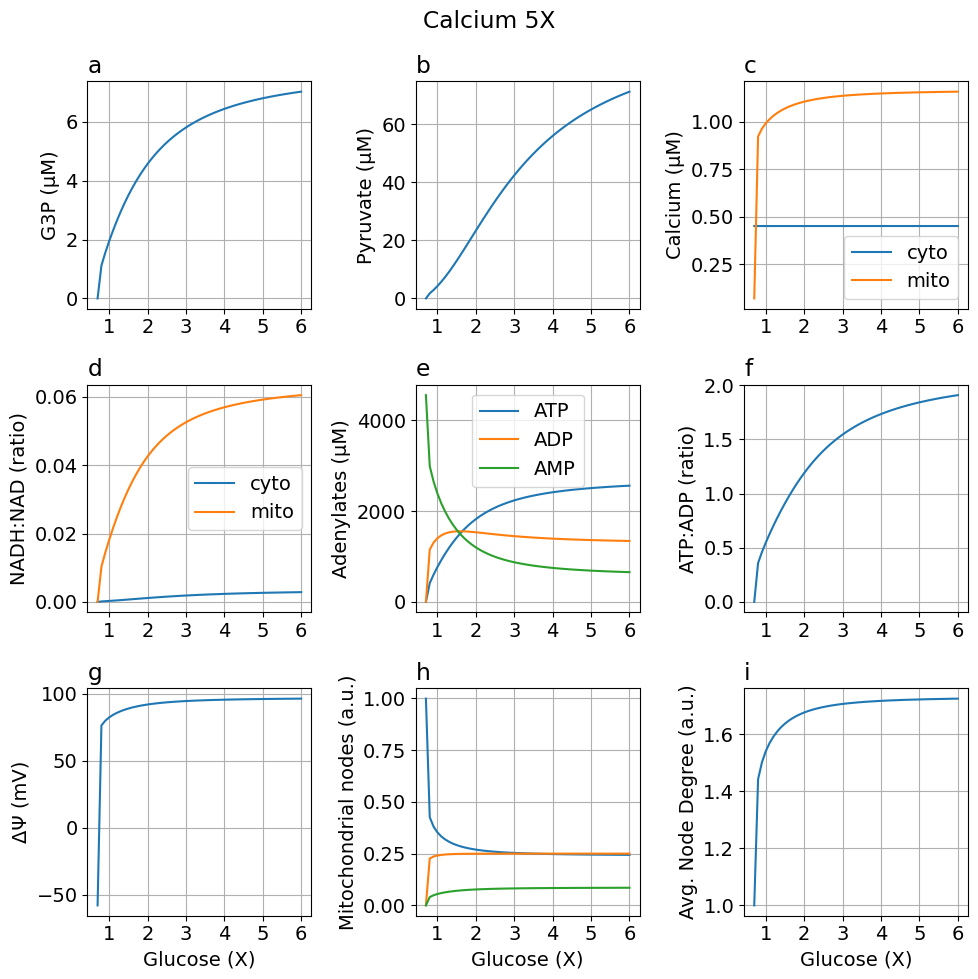
High calcium (5X)
fig_ca5 = plot_steady_state(glc, sim_ca5, sys, title="Calcium 5X")
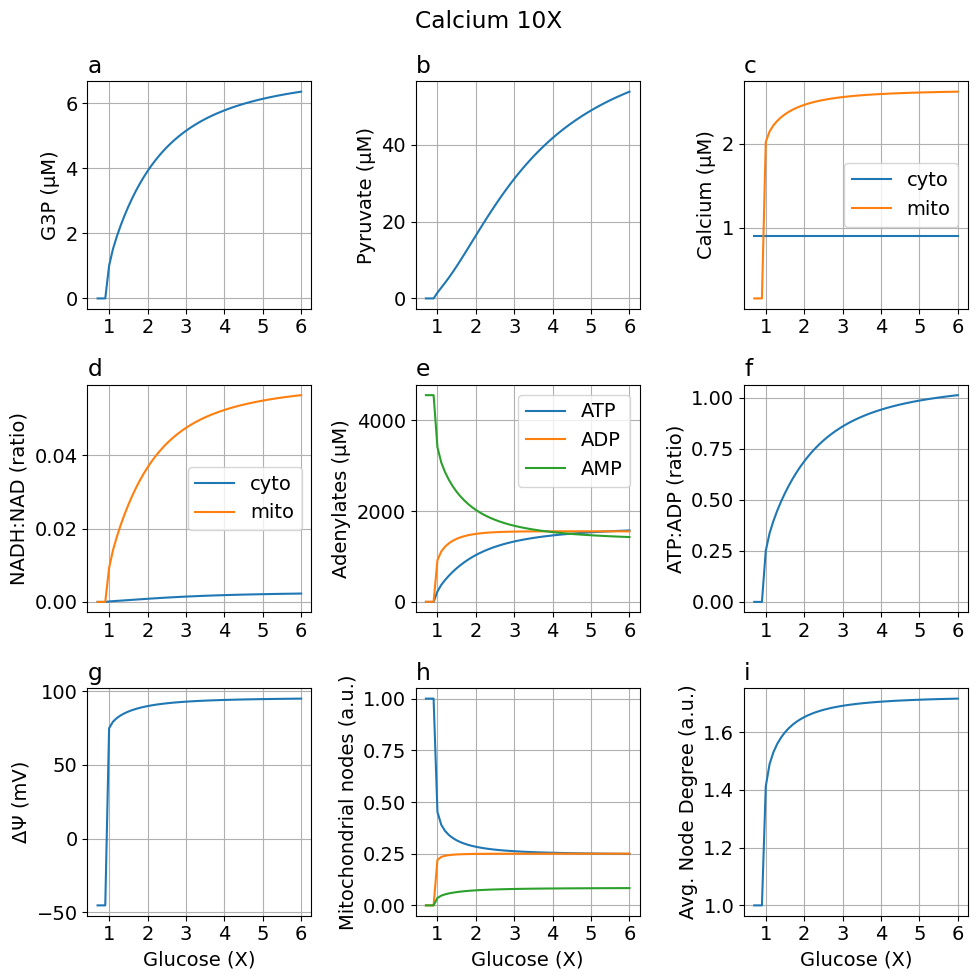
High calcium (10X)
fig_ca10 = plot_steady_state(glc, sim_ca10, sys, title="Calcium 10X")
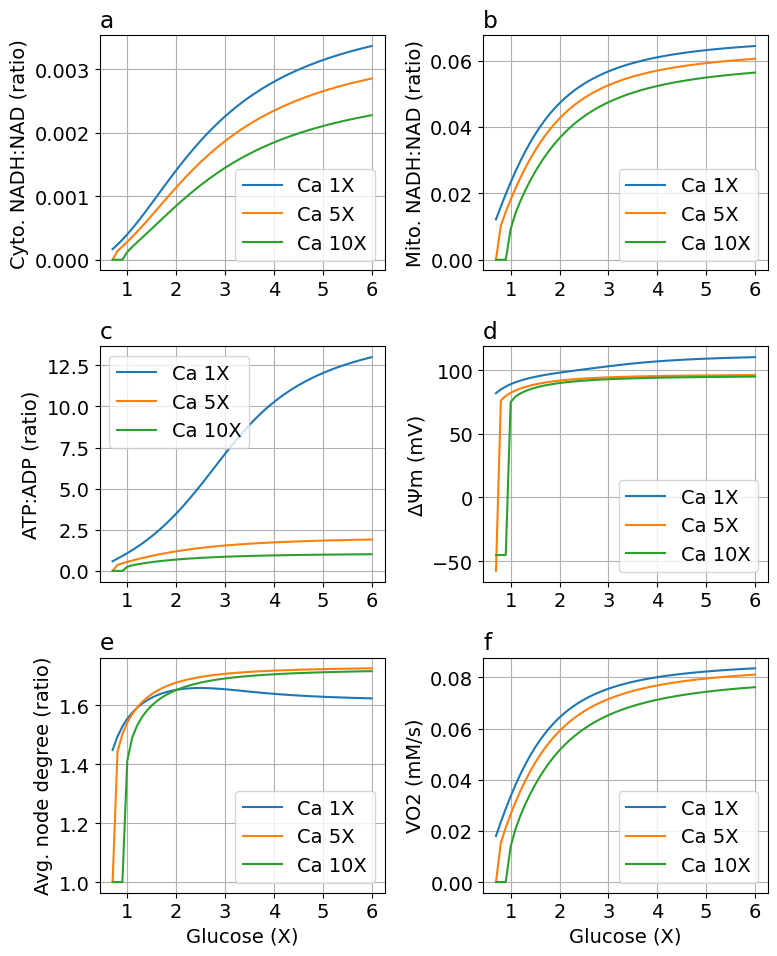
Comparing default and high calcium models#
function plot_comparision(glc, sim, sim_ca5, sim_ca10, sys;
figsize=(8, 10), title="", labels=["Ca 1X", "Ca 5X", "Ca 10X"]
)
@unpack G3P, Pyr, Ca_c, Ca_m, NADH_c, NADH_m, NAD_c, NAD_m, ATP_c, ADP_c, AMP_c, ΔΨm, degavg, J_O2 = sys
glc5 = glc ./ 5
numrows = 3
numcols = 2
fig, ax = plt.subplots(numrows, numcols; figsize)
ax[0, 0].set_title("a", loc="left")
ax[0, 0].set_ylabel("Cyto. NADH:NAD (ratio)")
k = NADH_c/NAD_c
yy = [extract(sim, k) extract(sim_ca5, k) extract(sim_ca10, k)]
lines = ax[0, 0].plot(glc5, yy)
ax[0, 0].legend(lines, labels)
ax[0, 1].set_title("b", loc="left")
ax[0, 1].set_ylabel("Mito. NADH:NAD (ratio)")
k = NADH_m/NAD_m
yy = [extract(sim, k) extract(sim_ca5, k) extract(sim_ca10, k)]
lines = ax[0, 1].plot(glc5, yy)
ax[0, 1].legend(lines, labels)
ax[1, 0].set_title("c", loc="left")
ax[1, 0].set_ylabel("ATP:ADP (ratio)")
k = ATP_c/ADP_c
yy = [extract(sim, k) extract(sim_ca5, k) extract(sim_ca10, k)]
lines = ax[1, 0].plot(glc5, yy)
ax[1, 0].legend(lines, labels)
ax[1, 1].set_title("d", loc="left")
ax[1, 1].set_ylabel("ΔΨm (mV)")
k = ΔΨm * 1000
yy = [extract(sim, k) extract(sim_ca5, k) extract(sim_ca10, k)]
lines = ax[1, 1].plot(glc5, yy)
ax[1, 1].legend(lines, labels)
ax[2, 0].set_title("e", loc="left")
ax[2, 0].set_ylabel("Avg. node degree (ratio)")
k = degavg
yy = [extract(sim, k) extract(sim_ca5, k) extract(sim_ca10, k)]
lines = ax[2, 0].plot(glc5, yy)
ax[2, 0].legend(lines, labels, loc="lower right")
ax[2, 0].set(xlabel="Glucose (X)")
ax[2, 1].set_title("f", loc="left")
ax[2, 1].set_ylabel("VO2 (mM/s)")
k = J_O2
yy = [extract(sim, k) extract(sim_ca5, k) extract(sim_ca10, k)]
lines = ax[2, 1].plot(glc5, yy)
ax[2, 1].legend(lines, labels)
ax[2, 1].set(xlabel="Glucose (X)")
for i in 0:numrows-1, j in 0:numcols-1
ax[i, j].set_xticks(1:6)
ax[i, j].grid()
end
fig.suptitle(title)
fig.tight_layout()
return fig
end
figcomp = plot_comparision(glc, sim, sim_ca5, sim_ca10, sys)
Export figure
exportTIF(figcomp, "S1_HighCa.tif")
Python: None
MMP vs #
function plot_dpsi_k(sim, sim_ca5, sim_ca10, sys; figsize=(6,6), title="", labels=["Ca 1X", "Ca 5X", "Ca 10X"])
@unpack ΔΨm, degavg = sys
fig, ax = plt.subplots(1, 1; figsize)
ax.plot(extract(sim, ΔΨm * 1000), extract(sim, degavg), "v", label=labels[1])
ax.plot(extract(sim_ca5, ΔΨm * 1000), extract(sim_ca5, degavg), "o", label=labels[2])
ax.plot(extract(sim_ca10, ΔΨm * 1000), extract(sim_ca10, degavg), "x", label=labels[3])
ax.set(xlabel="ΔΨm (mV)", ylabel="Average node degree", title=title)
ax.legend()
ax.grid()
return fig
end
fig = plot_dpsi_k(sim, sim_ca5, sim_ca10, sys)
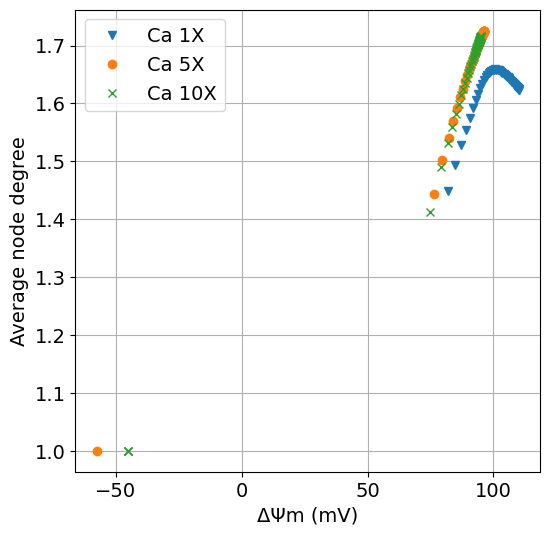
# exportTIF(fig, "S1_HighCa_dpsi_k.tif")
x-axis as Ca2+ and y-axis as average node degree#
function plot_ca_k(sim, sim_ca5, sim_ca10, sys; figsize=(6,6), title="", labels=["Ca 1X", "Ca 5X", "Ca 10X"])
@unpack Ca_m, degavg = sys
fig, ax = plt.subplots(1, 1; figsize)
ax.plot(extract(sim, Ca_m * 1000), extract(sim, degavg), "v", label=labels[1])
ax.plot(extract(sim_ca5, Ca_m * 1000), extract(sim_ca5, degavg), "o", label=labels[2])
ax.plot(extract(sim_ca10, Ca_m * 1000), extract(sim_ca10, degavg), "x", label=labels[3])
ax.set(xlabel="Mitochondrial Ca (mM)", ylabel="Average node degree", title=title)
ax.legend()
ax.grid()
return fig
end
fig = plot_ca_k(sim, sim_ca5, sim_ca10, sys)
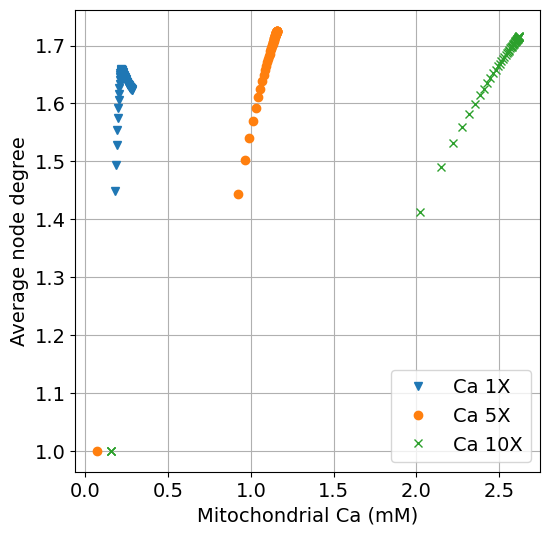
exportTIF(fig, "S1_HighCa_ca_k.tif")
Python: None
x-axis as ATP and y-axis as average node degree#
function plot_atp_k(sim, sim_ca5, sim_ca10, sys; figsize=(6,6), title="", labels=["Ca 1X", "Ca 5X", "Ca 10X"])
@unpack ATP_c, ADP_c, degavg = sys
k = ATP_c / ADP_c
fig, ax = plt.subplots(1, 1; figsize)
ax.plot(extract(sim, k), extract(sim, degavg), "v", label=labels[1])
ax.plot(extract(sim_ca5, k), extract(sim_ca5, degavg), "o", label=labels[2])
ax.plot(extract(sim_ca10, k), extract(sim_ca10, degavg), "x", label=labels[3])
ax.set(xlabel="ATP:ADP ratio", ylabel="Average node degree", title=title)
ax.legend()
ax.grid()
return fig
end
fig = plot_atp_k(sim, sim_ca5, sim_ca10, sys)
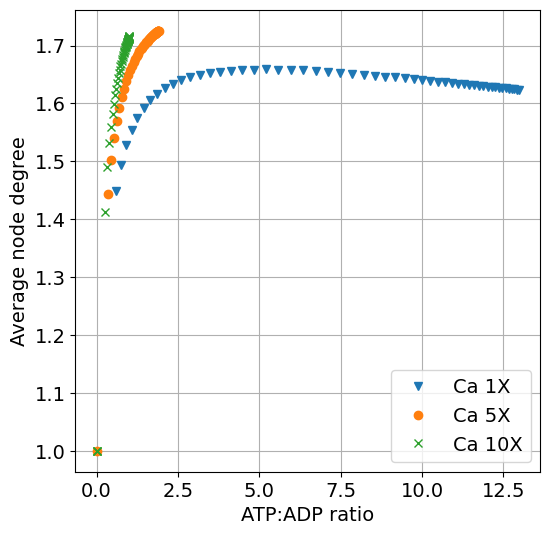
exportTIF(fig, "S1_HighCa_atp_k.tif")
Python: None
This notebook was generated using Literate.jl.

