Diabetic step response#
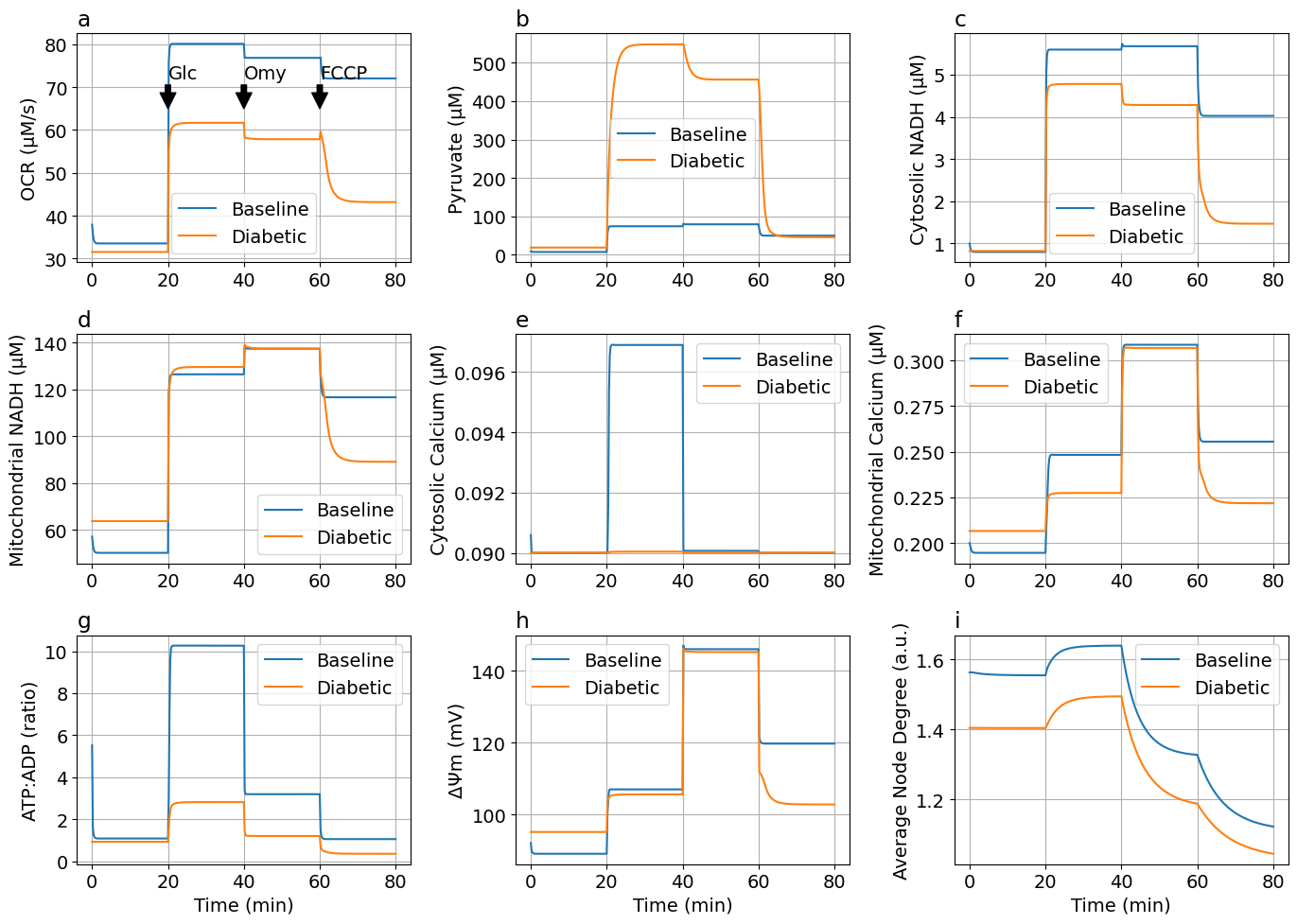
Glucose-Oligomycin-FCCP protocol#
Step responses to both glucose stimulation and chemical agents (I)
using OrdinaryDiffEq
using SteadyStateDiffEq
using DiffEqCallbacks
using ModelingToolkit
using MitochondrialDynamics
using MitochondrialDynamics: second, μM, mV, mM, Hz, minute
import PythonPlot as plt
plt.matplotlib.rcParams["font.size"] = 14
14
@named sys = make_model()
@unpack Glc, rPDH, rETC, rHL, rF1 = sys
tend = 80minute
ts = range(0, tend, 401)
tspan = (ts[begin], ts[end])
alg = TRBDF2()
prob = ODEProblem(sys, [], tspan)
prob_dm = ODEProblem(sys, [], tspan, [rPDH=>0.5, rETC=>0.75, rHL=>1.4, rF1=>0.5])
ssprob_dm = SteadyStateProblem(prob_dm)
sssol_dm = solve(ssprob_dm, DynamicSS(Rodas5()))
┌ Warning: `SciMLBase.ODEProblem(sys, u0, tspan, p; kw...)` is deprecated. Use
│ `SciMLBase.ODEProblem(sys, merge(if isempty(u0)
│ Dict()
│ else
│ Dict(unknowns(sys) .=> u0)
│ end, Dict(p)), tspan)` instead.
└ @ ModelingToolkit ~/.julia/packages/ModelingToolkit/b28X4/src/deprecations.jl:45
retcode: Success
u: 9-element Vector{Float64}:
0.03472910521628282
0.21820517998412747
0.4850068481164586
0.018234288180978456
0.0023800558865970513
0.09508018068076517
0.00020664324833474275
0.0008233256424150778
0.06366081984790718
Define events
function add_glucose!(i)
i.ps[Glc] = 20mM
end
add_glucose_cb = PresetTimeCallback(20minute, add_glucose!)
function add_oligomycin!(i)
i.ps[rF1] *= 0.1
end
add_oligomycin_cb = PresetTimeCallback(40minute, add_oligomycin!)
function add_rotenone!(i)
i.ps[rETC] *= 0.1
end
add_rotenone_cb = PresetTimeCallback(60minute, add_rotenone!)
function add_fccp!(i)
i.ps[rHL] *= 5
end
add_fccp_cb = PresetTimeCallback(60minute, add_fccp!)
prob_dm = remake(prob_dm, u0=sssol_dm.u)
cbs = CallbackSet(add_glucose_cb, add_oligomycin_cb, add_fccp_cb)
sols3 = solve(prob, alg; callback=cbs, saveat=ts)
solDMs3 = solve(prob_dm, alg; callback=cbs, saveat=ts);
function plot_figs2(sol, solDM; figsize=(14, 10), labels=["Baseline", "Diabetic"], jo2_mul=t->1, laststep="FCCP")
@unpack G3P, Pyr, NADH_c, NADH_m, Ca_c, Ca_m, ATP_c, ADP_c, ΔΨm, degavg, J_O2 = sol.prob.f.sys
ts = sol.t
tsm = ts ./ 60
jo2 = sol[J_O2 * 1000] .* jo2_mul.(ts)
pyr = sol[Pyr * 1000]
nadh_c = sol[NADH_c * 1000]
nadh_m = sol[NADH_m * 1000]
ca_c = sol[Ca_c * 1000]
ca_m = sol[Ca_m * 1000]
td = sol[ATP_c/ADP_c]
dpsi = sol[ΔΨm * 1000]
k = sol[degavg]
pyrDM = solDM[Pyr * 1000]
jo2DM = solDM[J_O2 * 1000] .* jo2_mul.(ts)
nadh_cDM = solDM[NADH_c * 1000]
nadh_mDM = solDM[NADH_m * 1000]
ca_cDM = solDM[Ca_c * 1000]
ca_mDM = solDM[Ca_m * 1000]
tdDM = solDM[ATP_c/ADP_c]
dpsiDM = solDM[ΔΨm * 1000]
kDM = solDM[degavg]
numrows = 3
numcols = 3
fig, ax = plt.subplots(numrows, numcols; figsize)
ax[0, 0].plot(tsm, jo2, label=labels[1])
ax[0, 0].plot(tsm, jo2DM, label=labels[2])
ax[0, 0].set(ylabel="OCR (μM/s)")
ax[0, 0].set_title("a", loc="left")
ax[0, 0].legend(loc="lower right")
ax[0, 0].annotate("Glc", xy=(20, 65), xytext=(20, 72),
arrowprops=Dict("shrink"=>3, "facecolor"=>"black"))
ax[0, 0].annotate("Omy", xy=(40, 65), xytext=(40, 72),
arrowprops=Dict("shrink"=>3, "facecolor"=>"black"))
ax[0, 0].annotate(laststep, xy=(60, 65), xytext=(60, 72),
arrowprops=Dict("shrink"=>3, "facecolor"=>"black"))
ax[0, 1].plot(tsm, pyr, label=labels[1])
ax[0, 1].plot(tsm, pyrDM, label=labels[2])
ax[0, 1].set(ylabel="Pyruvate (μM)")
ax[0, 1].set_title("b", loc="left")
ax[0, 2].plot(tsm, nadh_c, label=labels[1])
ax[0, 2].plot(tsm, nadh_cDM, label=labels[2])
ax[0, 2].set(ylabel="Cytosolic NADH (μM)")
ax[0, 2].set_title("c", loc="left")
ax[1, 0].plot(tsm, nadh_m, label=labels[1])
ax[1, 0].plot(tsm, nadh_mDM, label=labels[2])
ax[1, 0].set(ylabel="Mitochondrial NADH (μM)")
ax[1, 0].set_title("d", loc="left")
ax[1, 1].plot(tsm, ca_c, label=labels[1])
ax[1, 1].plot(tsm, ca_cDM, label=labels[2])
ax[1, 1].set(ylabel="Cytosolic Calcium (μM)")
ax[1, 1].set_title("e", loc="left")
ax[1, 2].plot(tsm, ca_m, label=labels[1])
ax[1, 2].plot(tsm, ca_mDM, label=labels[2])
ax[1, 2].set(ylabel="Mitochondrial Calcium (μM)")
ax[1, 2].set_title("f", loc="left")
ax[2, 0].plot(tsm, td, label=labels[1])
ax[2, 0].plot(tsm, tdDM, label=labels[2])
ax[2, 0].set(ylabel="ATP:ADP (ratio)", xlabel="Time (min)")
ax[2, 0].set_title("g", loc="left")
ax[2, 1].plot(tsm, dpsi, label=labels[1])
ax[2, 1].plot(tsm, dpsiDM, label=labels[2])
ax[2, 1].set(ylabel="ΔΨm (mV)", xlabel="Time (min)")
ax[2, 1].set_title("h", loc="left")
ax[2, 2].plot(tsm, k, label=labels[1])
ax[2, 2].plot(tsm, kDM, label=labels[2])
ax[2, 2].set(ylabel="Average Node Degree (a.u.)", xlabel="Time (min)")
ax[2, 2].set_title("i", loc="left")
for i in 0:numrows-1, j in 0:numcols-1
ax[i, j].legend()
ax[i, j].grid()
end
fig.tight_layout()
return fig
end
plot_figs2 (generic function with 1 method)
figs3 = plot_figs2(sols3, solDMs3)
TIFF file
exportTIF(figs3, "FigDM-Glucose-Oligomycin-FCCP.tif")
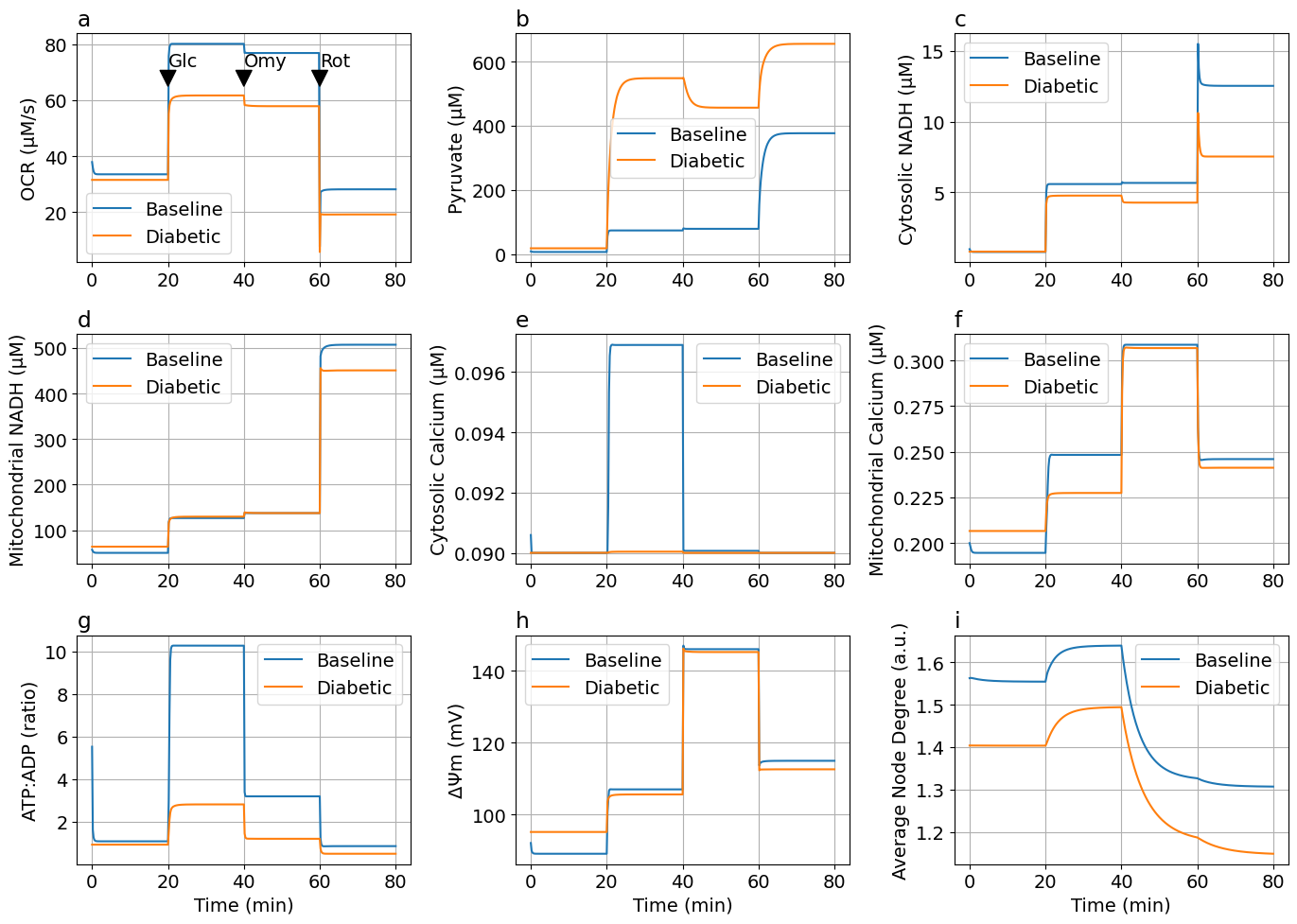
# Glucose-Oligomycin-Rotenone
prob5 = ODEProblem(sys, [], tspan)
prob_dm5 = ODEProblem(sys, sssol_dm.u, tspan, [rPDH=>0.5, rETC=>0.75, rHL=>1.4, rF1=>0.5])
cbs = CallbackSet(add_glucose_cb, add_oligomycin_cb, add_rotenone_cb)
sols5 = solve(prob5, alg; callback=cbs, saveat=ts)
sols5DM = solve(prob_dm5, alg; callback=cbs, saveat=ts)
jo2_mul = t -> 10 - 9 * (t>=60minute)
figs5 = plot_figs2(sols5, sols5DM; jo2_mul, laststep="Rot")
┌ Warning: `SciMLBase.ODEProblem(sys, u0, tspan, p; kw...)` is deprecated. Use
│ `SciMLBase.ODEProblem(sys, merge(if isempty(u0)
│ Dict()
│ else
│ Dict(unknowns(sys) .=> u0)
│ end, Dict(p)), tspan)` instead.
└ @ ModelingToolkit ~/.julia/packages/ModelingToolkit/b28X4/src/deprecations.jl:45
exportTIF(figs5, "FigDM-Glucose-Oligomycin-RotAA.tif")
Python: None
This notebook was generated using Literate.jl.

