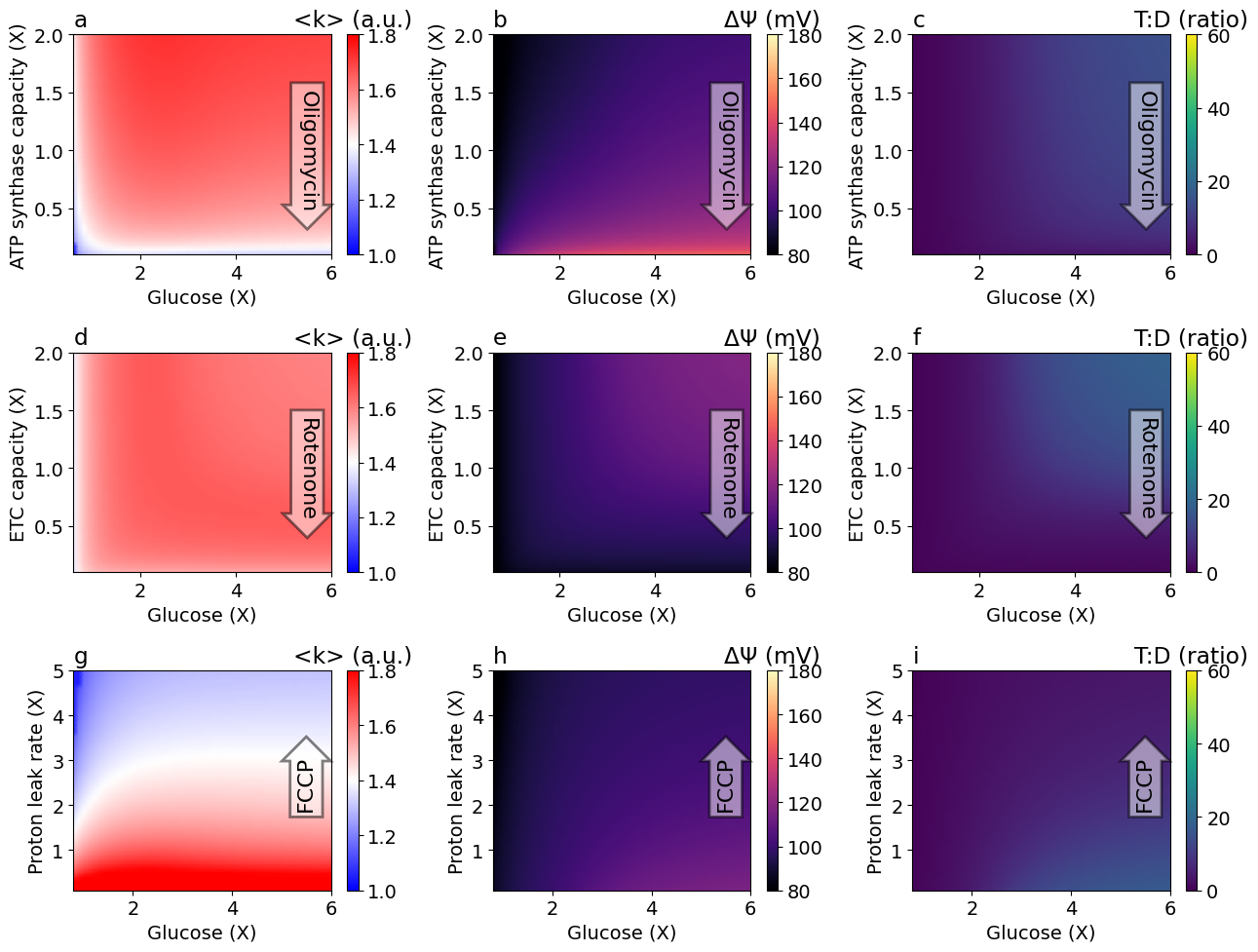
Figure 2#
Steady-state solutions for a range of glucose concentrations and OXPHOS capacities by chemicals.
using OrdinaryDiffEq
using SteadyStateDiffEq
using ModelingToolkit
using MitochondrialDynamics
import PythonPlot as plt
plt.matplotlib.rcParams["font.size"] = 14
14
@named sys = make_model()
@unpack Glc, rETC, rHL, rF1, rPDH = sys
prob = SteadyStateProblem(sys, [])
SteadyStateProblem with uType Vector{Float64}. In-place: true
u0: 9-element Vector{Float64}:
0.06
0.24
0.9
0.0087
0.0029
0.092
0.0002
0.001
0.057
Range for two parameters
rGlcF1 = range(3.0, 30.0, 51)
rGlcETC = range(3.0, 30.0, 51)
rGlcHL = range(4.0, 30.0, 51)
rf1 = range(0.1, 2.0, 51)
retc = range(0.1, 2.0, 51)
rhl = range(0.1, 5.0, 51)
0.1:0.098:5.0
2D steady states
function solve_fig3(glc, r, k, prob, alg=DynamicSS(Rodas5()))
newprob = remake(prob, p = [Glc=> glc, k => r])
return solve(newprob, alg)
end
solsf1 = [solve_fig3(glc, r, rF1, prob) for r in rf1, glc in rGlcF1];
solsetc = [solve_fig3(glc, r, rETC, prob) for r in retc, glc in rGlcETC];
solshl = [solve_fig3(glc, r, rHL, prob) for r in rhl, glc in rGlcHL];
function plot_fig3(;
figsize=(10, 10),
cmaps=["bwr", "magma", "viridis"],
ylabels=[
"ATP synthase capacity (X)",
"ETC capacity (X)",
"Proton leak rate (X)"
],
cbarlabels=["<k> (a.u.)", "ΔΨ (mV)", "T:D (ratio)"],
xxs=(rGlcF1, rGlcETC, rGlcHL),
xscale=5.0,
yys=(rf1, retc, rhl),
zs=(solsf1, solsetc, solshl),
extremes=((1.0, 1.8), (80.0, 180.0), (0.0, 60.0))
)
# mapping functions
@unpack degavg, ΔΨm, ATP_c, ADP_c = sys
fs = (s -> s[degavg], s -> s[ΔΨm * 1000] , s -> s[ATP_c / ADP_c])
fig, axes = plt.subplots(3, 3; figsize)
subtitle = [
"a" "b" "c";
"d" "e" "f";
"g" "h" "i";
]
for col in 1:3
f = fs[col]
cm = cmaps[col]
cbl = cbarlabels[col]
vmin, vmax = extremes[col]
# lvls = LinRange(vmin, vmax, levels)
for row in 1:3
xx = xxs[row] ./ xscale
yy = yys[row]
z = zs[row]
ax = axes[row-1, col-1]
ylabel = ylabels[row]
mesh = ax.pcolormesh(
xx, yy, map(f, z);
shading="gouraud",
rasterized=true,
cmap=cm,
vmin=vmin,
vmax=vmax
)
ax.set(ylabel=ylabel, xlabel="Glucose (X)")
ax.set_title(subtitle[row, col], loc="left")
# Arrow annotation: https://matplotlib.org/stable/tutorials/text/annotations.html#plotting-guide-annotation
if row == 1
ax.text(5.5, 1, "Oligomycin", ha="center", va="center", rotation=-90, size=16, bbox=Dict("boxstyle" => "rarrow", "fc" => "w", "ec" => "k", "lw" => 2, "alpha" => 0.5))
elseif row == 2
ax.text(5.5, 1, "Rotenone", ha="center", va="center", rotation=-90, size=16, bbox=Dict("boxstyle" => "rarrow", "fc" => "w", "ec" => "k", "lw" => 2, "alpha" => 0.5))
elseif row == 3
ax.text(5.5, 2.5, "FCCP", ha="center", va="center", rotation=90, size=16, bbox=Dict("boxstyle" => "rarrow", "fc" => "w", "ec" => "k", "lw" => 2, "alpha" => 0.5))
end
cbar = fig.colorbar(mesh, ax=ax)
cbar.ax.set_title(cbl)
end
end
fig.tight_layout()
return fig
end
plot_fig3 (generic function with 1 method)
fig = plot_fig3(figsize=(13, 10))
Export figure
exportTIF(fig, "Fig2-2Dsteadystate.tif")
Python: None
This notebook was generated using Literate.jl.

