using ModelingToolkit
using OrdinaryDiffEq, SteadyStateDiffEq, DiffEqCallbacks
using Plots
using CSV
using DataFrames
using CaMKIIModel
using CaMKIIModel: second
Plots.default(lw=1.5)Pacing data
Experiments vs simulations.
sys = build_neonatal_ecc_sys(simplify=true, reduce_iso=true, reduce_camk=true)
tend = 500second
prob = ODEProblem(sys, [], tend)
stimstart = 100second
stimend = 300second
@unpack Istim = sys
alg = KenCarp47()KenCarp47(; linsolve = nothing, nlsolve = OrdinaryDiffEqNonlinearSolve.NLNewton{Rational{Int64}, Rational{Int64}, Rational{Int64}, Nothing}(1//100, 10, 1//5, 1//5, false, true, nothing), precs = DEFAULT_PRECS, smooth_est = true, extrapolant = linear, controller = PI, autodiff = ADTypes.AutoForwardDiff(),)Pacing duration and CaMKII activity
Experiments
durationdf = CSV.read(joinpath(@__DIR__, "data/CaMKAR-duration.csv"), DataFrame)
ts = durationdf[!, "Time(sec)"]
fifteen = durationdf[!, "1Hz 15sec (Mean)"]
fifteen_error = durationdf[!, "1Hz 15sec (SD)"] ./ sqrt.(durationdf[!, "1Hz 15sec (N)"])
thirty = durationdf[!, "1Hz 30sec (Mean)"] .+ 0.25
thirty_error = durationdf[!, "1Hz 30sec (SD)"] ./ sqrt.(durationdf[!, "1Hz 30sec (N)"])
sixty = durationdf[!, "1Hz 60sec (Mean)"]
sixty_error = durationdf[!, "1Hz 60sec (SD)"] ./ sqrt.(durationdf[!, "1Hz 60sec (N)"])
ninety = durationdf[!, "1Hz 90sec (Mean)"] .- 0.25
ninety_error = durationdf[!, "1Hz 90sec (SD)"] ./ sqrt.(durationdf[!, "1Hz 90sec (N)"])
# 30 sec timeseries +0.25 and 90 sec timeseries -0.25 for a consistent baseline before pacing.
plot(ts, fifteen, yerr=fifteen_error, lab="15 sec", color=:blue, markerstrokecolor=:blue)
plot!(ts, thirty, yerr=thirty_error, lab="30 sec", color=:red, markerstrokecolor=:red)
plot!(ts, sixty, yerr=sixty_error, lab="60 sec", color=:orange, markerstrokecolor=:orange)
plot!(ts, ninety, yerr=ninety_error, lab="90 sec", color=:green, markerstrokecolor=:green)
plot!(title="Experiment", xlabel="Time (s)", ylabel="CaMKII activity (AU)")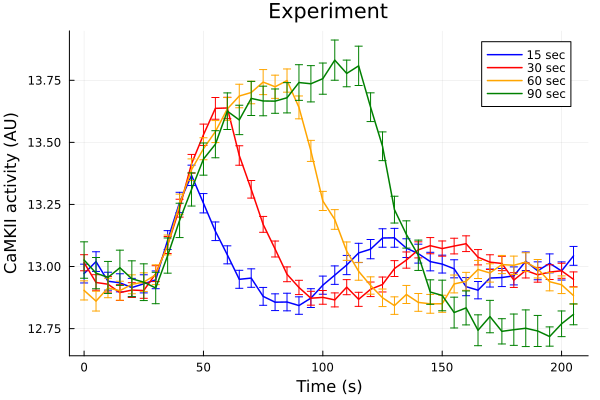
savefig("pacing-duration-exp.pdf")"/home/runner/work/camkii-cardiomyocyte-model/camkii-cardiomyocyte-model/docs/pacing-duration-exp.pdf"Simulation
stimstart = 30second
callback15 = build_stim_callbacks(Istim, stimstart + 15second; period=1second, starttime=stimstart)
sol15 = solve(prob, alg; callback=callback15)
callback30 = build_stim_callbacks(Istim, stimstart + 30second; period=1second, starttime=stimstart)
sol30 = solve(prob, alg; callback=callback30)
callback60 = build_stim_callbacks(Istim, stimstart + 60second; period=1second, starttime=stimstart)
sol60 = solve(prob, alg; callback=callback60)
callback90 = build_stim_callbacks(Istim, stimstart + 90second; period=1second, starttime=stimstart)
sol90 = solve(prob, alg; callback=callback90)
idxs = (sys.t / 1000, sys.CaMKAct * 100)
plot(sol15, idxs=idxs, tspan=(0second, 205second), lab="15 sec", color=:blue)
plot!(sol30, idxs=idxs, tspan=(0second, 205second), lab="30 sec", color=:red)
plot!(sol60, idxs=idxs, tspan=(0second, 205second), lab="60 sec", color=:orange)
plot!(sol90, idxs=idxs, tspan=(0second, 205second), lab="90 sec", color=:green)
plot!(title="Simulation", xlabel="Time (s)", ylabel="CaMKII activity (%)")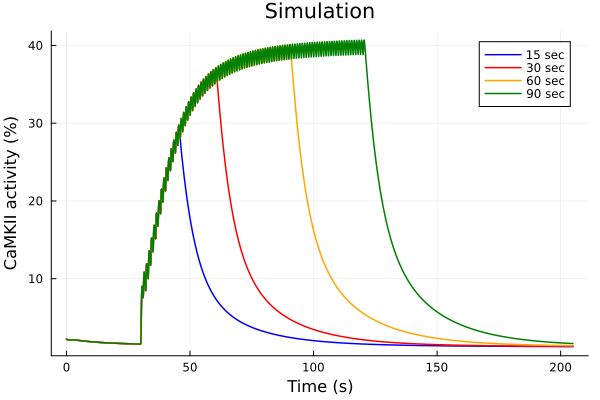
savefig("pacing-duration-sim.pdf")"/home/runner/work/camkii-cardiomyocyte-model/camkii-cardiomyocyte-model/docs/pacing-duration-sim.pdf"Phosphorylated fraction
idxs = (sys.t / 1000, (sys.CaMKP + sys.CaMKA + sys.CaMKA2) * 100)
plot(sol15, idxs=idxs, tspan=(0second, 205second), lab="15 sec", color=:blue)
plot!(sol30, idxs=idxs, tspan=(0second, 205second), lab="30 sec", color=:red)
plot!(sol60, idxs=idxs, tspan=(0second, 205second), lab="60 sec", color=:orange)
plot!(sol90, idxs=idxs, tspan=(0second, 205second), lab="90 sec", color=:green)
plot!(title="Simulation", xlabel="Time (s)", ylabel="Phosphorylated CaMKII (%)")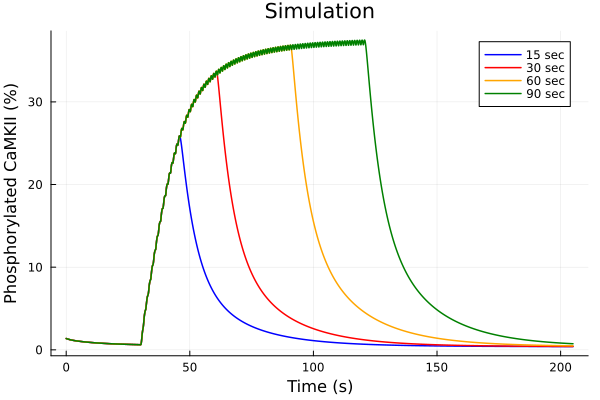
savefig("pacing-duration-phos.pdf")"/home/runner/work/camkii-cardiomyocyte-model/camkii-cardiomyocyte-model/docs/pacing-duration-phos.pdf"Pacing frequency and CaMKII activity
Experiments
freqdf = CSV.read(joinpath(@__DIR__, "data/CaMKAR-freq.csv"), DataFrame)
ts = 0:5:205
onehz = freqdf[!, "1Hz (Mean)"]
onehz_error = freqdf[!, "1Hz (SD)"] ./ sqrt.(freqdf[!, "1Hz (N)"])
twohz = freqdf[!, "2Hz (Mean)"]
twohz_error = freqdf[!, "2Hz (SD)"] ./ sqrt.(freqdf[!, "2Hz (N)"])
plot(ts, onehz, yerr=onehz_error, lab="1 Hz", color=:blue, markerstrokecolor=:blue)
plot!(ts, twohz, yerr=twohz_error, lab="2 Hz", color=:red, markerstrokecolor=:red)
plot!(title="Experiment", xlabel="Time (s)", ylabel="CaMKII activity (AU)")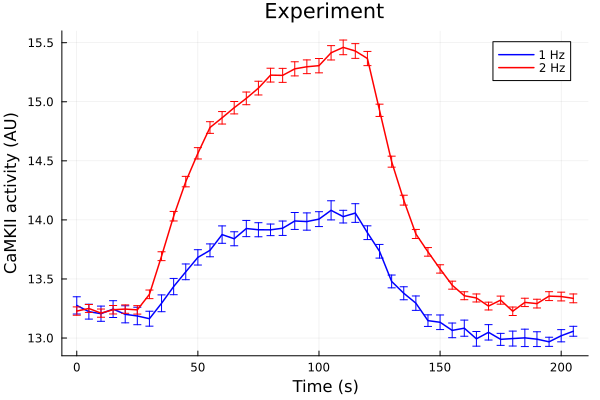
savefig("pacing-frequency-exp.pdf")"/home/runner/work/camkii-cardiomyocyte-model/camkii-cardiomyocyte-model/docs/pacing-frequency-exp.pdf"Simulations
tend = 205.0second
prob = ODEProblem(sys, [], tend)
stimstart = 30.0second
stimend = 120.0second
callback = build_stim_callbacks(Istim, stimend; period=1second, starttime=stimstart)
sol1 = solve(prob, alg; callback)
callback2 = build_stim_callbacks(Istim, stimend; period=0.5second, starttime=stimstart)
sol2 = solve(prob, alg; callback=callback2)
idxs = (sys.t / 1000, sys.CaMKAct * 100)
plot(sol1, idxs=idxs, lab="1 Hz", color=:blue)
plot!(sol2, idxs=idxs, lab="2 Hz", color=:red)
plot!(title="Simulation", xlabel="Time (s)", ylabel="CaMKII activity (%)")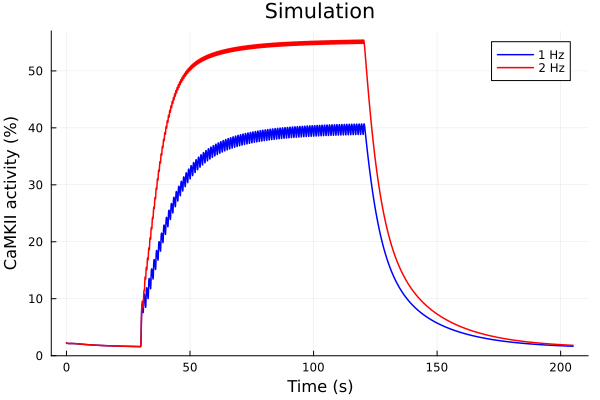
savefig("pacing-frequency-sim.pdf")"/home/runner/work/camkii-cardiomyocyte-model/camkii-cardiomyocyte-model/docs/pacing-frequency-sim.pdf"idxs = (sys.t / 1000, (sys.CaMKP + sys.CaMKA + sys.CaMKA2) * 100)
plot(sol1, idxs=idxs, lab="1 Hz", color=:blue)
plot!(sol2, idxs=idxs, lab="2 Hz", color=:red)
plot!(title="Simulation", xlabel="Time (s)", ylabel="Phosphorylated CaMKII (%)")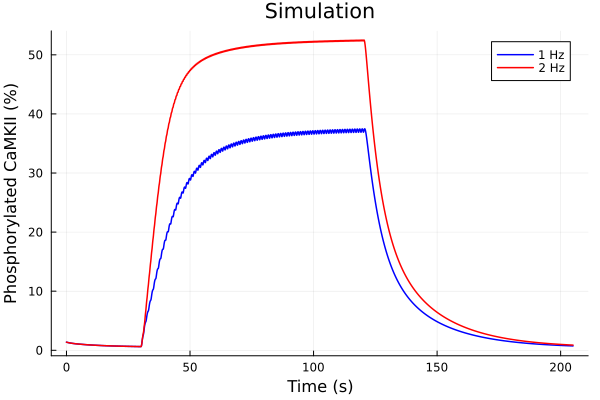
savefig("pacing-frequency-phos.pdf")"/home/runner/work/camkii-cardiomyocyte-model/camkii-cardiomyocyte-model/docs/pacing-frequency-phos.pdf"Runtime information
using InteractiveUtils
InteractiveUtils.versioninfo()Julia Version 1.12.4
Commit 01a2eadb047 (2026-01-06 16:56 UTC)
Build Info:
Official https://julialang.org release
Platform Info:
OS: Linux (x86_64-linux-gnu)
CPU: 4 × AMD EPYC 7763 64-Core Processor
WORD_SIZE: 64
LLVM: libLLVM-18.1.7 (ORCJIT, znver3)
GC: Built with stock GC
Threads: 4 default, 1 interactive, 4 GC (on 4 virtual cores)
Environment:
JULIA_CPU_TARGET = generic;icelake-server,clone_all;znver3,clone_all
JULIA_CONDAPKG_OFFLINE = true
JULIA_CONDAPKG_BACKEND = Null
JULIA_CI = true
LD_LIBRARY_PATH = /opt/hostedtoolcache/Python/3.14.2/x64/lib
JULIA_NUM_THREADS = autousing Pkg
Pkg.status()Project CaMKIIModel v0.7.0
Status `~/work/camkii-cardiomyocyte-model/camkii-cardiomyocyte-model/Project.toml`
[336ed68f] CSV v0.10.15
[a93c6f00] DataFrames v1.8.1
[459566f4] DiffEqCallbacks v4.11.0
[f6369f11] ForwardDiff v1.3.1
[682c06a0] JSON v1.3.0
[23fbe1c1] Latexify v0.16.10
[98b081ad] Literate v2.21.0
[2fda8390] LsqFit v0.15.1
⌅ [961ee093] ModelingToolkit v10.31.2
[77ba4419] NaNMath v1.1.3
[1dea7af3] OrdinaryDiffEq v6.105.0
[91a5bcdd] Plots v1.41.4
[2913bbd2] StatsBase v0.34.9
[9672c7b4] SteadyStateDiffEq v2.8.0
[ea8e919c] SHA v0.7.0
Info Packages marked with ⌅ have new versions available but compatibility constraints restrict them from upgrading. To see why use `status --outdated`This notebook was generated using Literate.jl.