Solving Ordinary Differential Equations (ODEs) in Python#
Source: Scipy’s documentation.
odeint(model,y0,t,args=(...))
import numpy as np
from scipy.integrate import solve_ivp
import matplotlib.pyplot as plt
%matplotlib inline
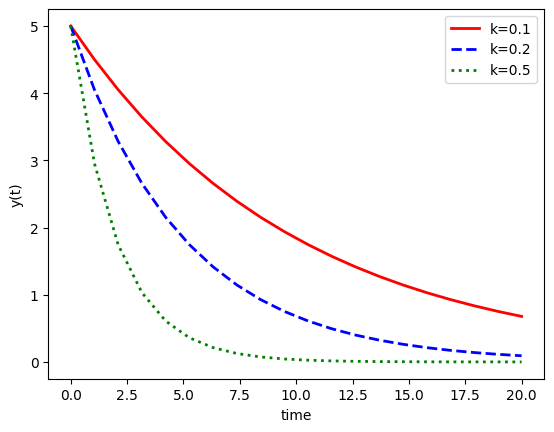
Exponential decays#
# function that returns dy/dt
def model(t, y, k):
dydt = -k * y[0]
return [dydt]
# initial condition
y0 = [5]
tspan = (0., 20.)
# solve ODEs
y1 = solve_ivp(model, tspan, y0, args=(0.1,), dense_output=True)
y2 = solve_ivp(model, tspan, y0, args=(0.2,), dense_output=True)
y3 = solve_ivp(model, tspan, y0, args=(0.5,), dense_output=True)
ts = np.linspace(tspan[0], tspan[1], 20)
# plot results
plt.figure()
plt.plot(ts, y1.sol(ts).T, 'r-',linewidth=2,label='k=0.1')
plt.plot(ts, y2.sol(ts).T, 'b--',linewidth=2,label='k=0.2')
plt.plot(ts, y3.sol(ts).T, 'g:',linewidth=2,label='k=0.5')
plt.xlabel('time')
plt.ylabel('y(t)')
plt.legend()
<matplotlib.legend.Legend at 0x7f4ef7f67b60>
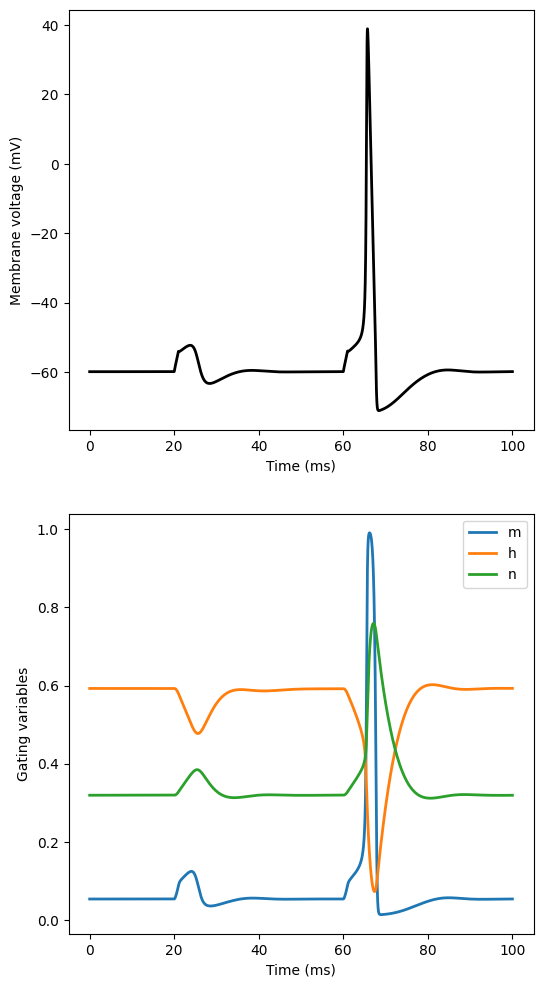
Hodgkin-Huxley electrophysiology model#
"""
Hodgkin-Huxley model of excitable barnacle muscle fiber
reviewed in Rinzel (1990) Bulletin of Mathematical Biology 52 pp. 5-23.
"""
from math import exp, expm1
def _iStim(t):
return (20 < t <= 21) * -6.8 + (60 < t <= 61) * -6.9
# Model
def hh_rhs(t, y, E_N, E_K, E_LEAK, G_N_BAR, G_K_BAR, G_LEAK, C_M):
# Equations
v, m, h, n = y
mAlfaV = -0.10 * (v + 35)
mAlfa = mAlfaV / expm1(mAlfaV)
mBeta = 4.0 * exp(-(v + 60) / 18.0)
dm = -(mAlfa + mBeta) * m + mAlfa
hAlfa = 0.07 * exp(-(v+60)/20)
hBeta = 1 / (exp(-(v+30)/10) + 1)
dh = -(hAlfa + hBeta) * h + hAlfa
iNa = G_N_BAR * (v - E_N) * (m**3) * h
nAlfaV = -0.1 * (v+50)
nAlfa = 0.1 * nAlfaV / expm1(nAlfaV)
nBeta = 0.125 * exp( -(v+60) / 80)
dn = -(nAlfa + nBeta) * n + nAlfa
iK = G_K_BAR * (v - E_K) * (n**4)
iLeak = G_LEAK * (v - E_LEAK)
iSt = _iStim(t)
dv = -(iNa + iK + iLeak + iSt) / C_M
return [dv, dm, dh, dn]
# Initial conditions
y0 = v, m, h, n = -59.8977, 0.0536, 0.5925, 0.3192
# Parameters
args = (
55.0, # Reversal potential of Na
-72.0, # Reversal potential of K
-49.0, # Reversal potential of leaky channels
120.0, # Max. Na channel conductance
36.0, # Max. K channel conductance
0.3, # Max. leak channel conductance
1.0 # membrane capacitance
)
# time span
tStart, tEnd = 0, 100
%%time
sol2 = solve_ivp(hh_rhs, (tStart, tEnd), y0, args=args, method='LSODA', rtol=1e-7, atol=1e-7)
CPU times: user 23.5 ms, sys: 31 μs, total: 23.5 ms
Wall time: 23.1 ms
# Plotting
fig, axs = plt.subplots(nrows=2, figsize=(6, 12))
axs[0].plot(sol2.t, sol2.y[0, :].T, 'k-', linewidth=2)
axs[0].set_xlabel("Time (ms)")
axs[0].set_ylabel("Membrane voltage (mV)")
axs[1].plot(sol2.t, sol2.y[1:4, :].T, linewidth=2, label=["m", "h", "n"])
axs[1].set_xlabel("Time (ms)")
axs[1].set_ylabel("Gating variables")
axs[1].legend()
<matplotlib.legend.Legend at 0x7f4ef7c53e00>