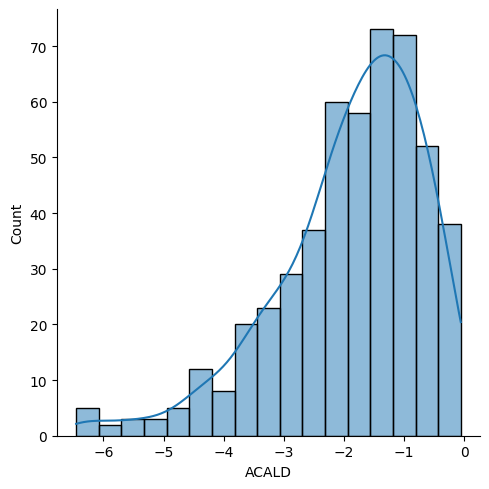
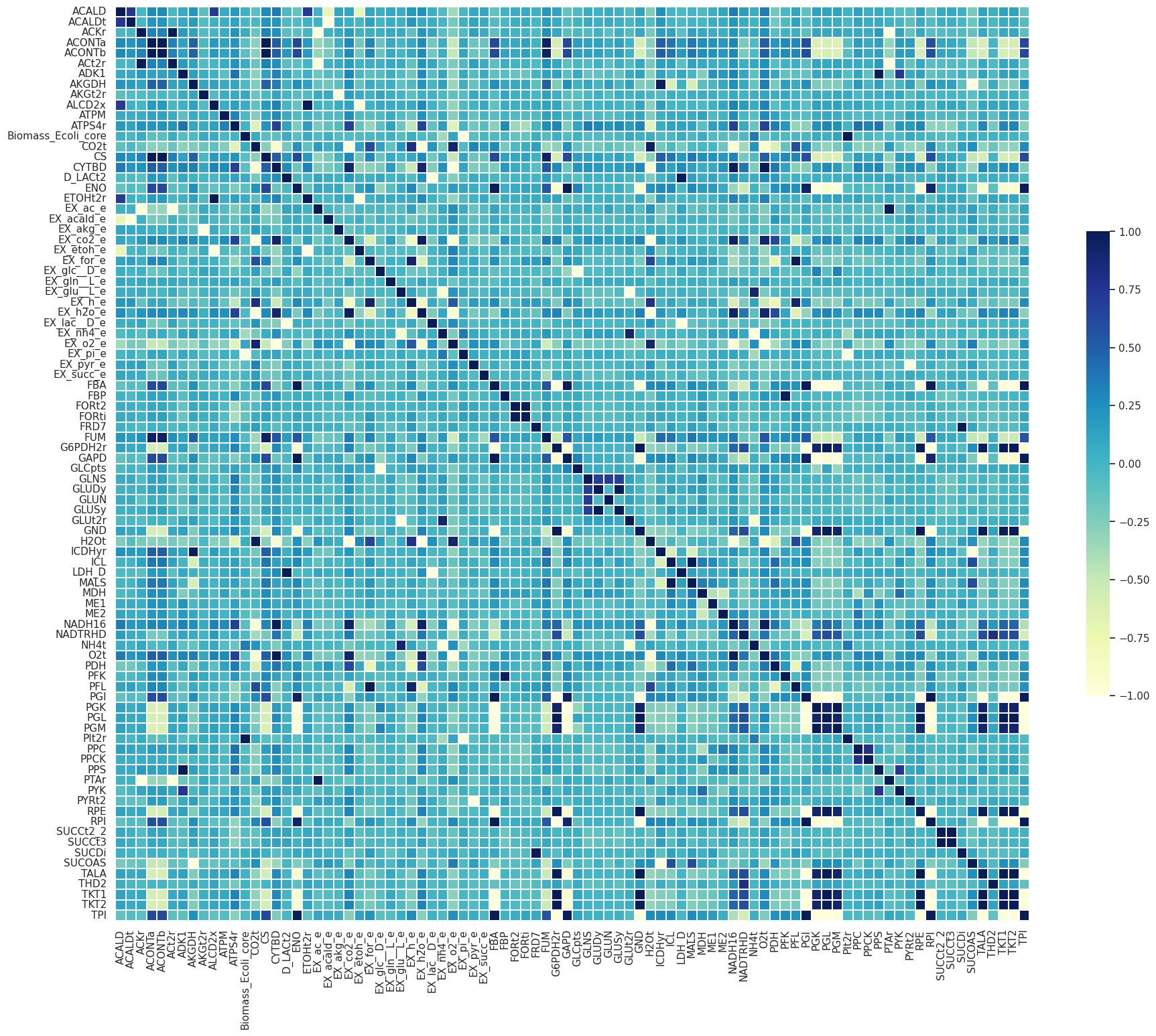
['DM_aacald_c',
'EX_14glucan_e',
'EX_23camp_e',
'EX_23ccmp_e',
'EX_23cgmp_e',
'EX_23cump_e',
'EX_23dappa_e',
'EX_26dap__M_e',
'EX_2ddglcn_e',
'EX_34dhpac_e',
'EX_3amp_e',
'EX_3cmp_e',
'EX_3gmp_e',
'EX_3hcinnm_e',
'EX_3hpppn_e',
'EX_3ump_e',
'EX_4hoxpacd_e',
'EX_LalaDglu_e',
'EX_LalaLglu_e',
'EX_acac_e',
'EX_acgal_e',
'EX_acgal1p_e',
'EX_acgam_e',
'EX_acgam1p_e',
'EX_acmana_e',
'EX_acmum_e',
'EX_acnam_e',
'EX_adocbl_e',
'EX_ag_e',
'EX_ala_B_e',
'EX_all__D_e',
'EX_amp_e',
'EX_arab__L_e',
'EX_arbt_e',
'EX_arbtn_e',
'EX_arbtn_fe3_e',
'EX_ascb__L_e',
'EX_aso3_e',
'EX_btn_e',
'EX_but_e',
'EX_butso3_e',
'EX_cbi_e',
'EX_cd2_e',
'EX_chol_e',
'EX_chtbs_e',
'EX_cm_e',
'EX_cmp_e',
'EX_cpgn_e',
'EX_cpgn_un_e',
'EX_crn_e',
'EX_crn__D_e',
'EX_csn_e',
'EX_cu_e',
'EX_cyan_e',
'EX_cynt_e',
'EX_cys__D_e',
'EX_dad_2_e',
'EX_damp_e',
'EX_dca_e',
'EX_dcmp_e',
'EX_dcyt_e',
'EX_ddca_e',
'EX_dgmp_e',
'EX_dgsn_e',
'EX_dimp_e',
'EX_din_e',
'EX_dms_e',
'EX_dmso_e',
'EX_dopa_e',
'EX_doxrbcn_e',
'EX_dtmp_e',
'EX_dump_e',
'EX_duri_e',
'EX_ethso3_e',
'EX_f6p_e',
'EX_fald_e',
'EX_fe3dcit_e',
'EX_fe3dhbzs_e',
'EX_fe3hox_e',
'EX_fe3hox_un_e',
'EX_fecrm_e',
'EX_fecrm_un_e',
'EX_feoxam_e',
'EX_feoxam_un_e',
'EX_fru_e',
'EX_frulys_e',
'EX_fruur_e',
'EX_fuc__L_e',
'EX_fum_e',
'EX_fusa_e',
'EX_g1p_e',
'EX_g3pc_e',
'EX_g3pi_e',
'EX_g3ps_e',
'EX_g6p_e',
'EX_gal_e',
'EX_gal_bD_e',
'EX_gal1p_e',
'EX_galct__D_e',
'EX_galctn__D_e',
'EX_galctn__L_e',
'EX_galt_e',
'EX_galur_e',
'EX_gam_e',
'EX_gam6p_e',
'EX_gbbtn_e',
'EX_gdp_e',
'EX_glcr_e',
'EX_glcur_e',
'EX_glcur1p_e',
'EX_gln__L_e',
'EX_glyb_e',
'EX_glyc2p_e',
'EX_gmp_e',
'EX_gsn_e',
'EX_gthox_e',
'EX_gtp_e',
'EX_h2_e',
'EX_h2o2_e',
'EX_hacolipa_e',
'EX_halipa_e',
'EX_hdca_e',
'EX_hdcea_e',
'EX_hg2_e',
'EX_imp_e',
'EX_inost_e',
'EX_isetac_e',
'EX_lcts_e',
'EX_lipoate_e',
'EX_lyx__L_e',
'EX_mal__D_e',
'EX_malt_e',
'EX_malthx_e',
'EX_maltpt_e',
'EX_malttr_e',
'EX_maltttr_e',
'EX_man_e',
'EX_man6p_e',
'EX_manglyc_e',
'EX_melib_e',
'EX_met__D_e',
'EX_met__L_e',
'EX_metsox_R__L_e',
'EX_metsox_S__L_e',
'EX_mincyc_e',
'EX_minohp_e',
'EX_mmet_e',
'EX_mnl_e',
'EX_mso3_e',
'EX_n2o_e',
'EX_na1_e',
'EX_nac_e',
'EX_nmn_e',
'EX_no_e',
'EX_no2_e',
'EX_no3_e',
'EX_novbcn_e',
'EX_o16a4colipa_e',
'EX_o2s_e',
'EX_ocdca_e',
'EX_ocdcea_e',
'EX_octa_e',
'EX_orot_e',
'EX_pacald_e',
'EX_peamn_e',
'EX_pnto__R_e',
'EX_ppa_e',
'EX_ppal_e',
'EX_pppn_e',
'EX_ppt_e',
'EX_progly_e',
'EX_psclys_e',
'EX_pser__L_e',
'EX_pydam_e',
'EX_pydx_e',
'EX_pydxn_e',
'EX_r5p_e',
'EX_rfamp_e',
'EX_rib__D_e',
'EX_rmn_e',
'EX_sbt__D_e',
'EX_sel_e',
'EX_ser__D_e',
'EX_skm_e',
'EX_slnt_e',
'EX_so2_e',
'EX_so3_e',
'EX_sucr_e',
'EX_sulfac_e',
'EX_tartr__D_e',
'EX_tartr__L_e',
'EX_taur_e',
'EX_tcynt_e',
'EX_thm_e',
'EX_thrp_e',
'EX_tma_e',
'EX_tmao_e',
'EX_tre_e',
'EX_tsul_e',
'EX_ttdca_e',
'EX_ttdcea_e',
'EX_ttrcyc_e',
'EX_tungs_e',
'EX_tym_e',
'EX_tyrp_e',
'EX_uacgam_e',
'EX_udpacgal_e',
'EX_udpg_e',
'EX_udpgal_e',
'EX_udpglcur_e',
'EX_ump_e',
'EX_xmp_e',
'EX_xyl__D_e',
'EX_xylu__L_e',
'14GLUCANabcpp',
'14GLUCANtexi',
'23CAMPtex',
'23CCMPtex',
'23CGMPtex',
'23CUMPtex',
'23DAPPAt2pp',
'23DAPPAtex',
'23PDE2pp',
'23PDE4pp',
'23PDE7pp',
'23PDE9pp',
'26DAHtex',
'2DGULRGx',
'2DGULRGy',
'2DGULRx',
'2DGULRy',
'34dhpactex',
'3AMPtex',
'3CMPtex',
'3GMPtex',
'3HCINNMH',
'3HPPPNH',
'3KGK',
'3NTD2pp',
'3NTD4pp',
'3NTD7pp',
'3NTD9pp',
'3OXCOAT',
'3UMPtex',
'42A12BOOXpp',
'4HOXPACDtex',
'4HTHRS',
'AAMYL',
'AAMYLpp',
'AB6PGH',
'ACACCT',
'ACACt2pp',
'ACACtex',
'ACANTHAT',
'ACBIPGT',
'ACGAL1PPpp',
'ACGAL1Ptex',
'ACGALtex',
'ACGAM1PPpp',
'ACGAM1Ptex',
'ACGAptspp',
'ACGAtex',
'ACMANAptspp',
'ACMANAtex',
'ACMUMptspp',
'ACMUMtex',
'ACNAMt2pp',
'ACNAMtex',
'ACNML',
'ACONIs',
'ACONMT',
'ACPS1',
'ADOCBIK',
'ADOCBLS',
'ADOCBLabcpp',
'ADOCBLtonex',
'AGt3',
'ALATRS',
'ALDD19xr',
'ALDD3y',
'ALDD4',
'ALLK',
'ALLPI',
'ALLULPE',
'ALLabcpp',
'ALLtex',
'ALPATE160pp',
'ALPATG160pp',
'ALTRH',
'AMALT1',
'AMALT2',
'AMALT3',
'AMALT4',
'AMANAPEr',
'AMANK',
'AMPTASEPG',
'AMPtex',
'AP5AH',
'APCS',
'APPLDHr',
'ARAI',
'ARBTptspp',
'ARBTtex',
'ARBtex',
'ARGTRS',
'ASCBPL',
'ASCBptspp',
'ASCBtex',
'ASNTRS',
'ASO3t8pp',
'ASO3tex',
'ASPTRS',
'ASR',
'BALAt2pp',
'BALAtex',
'BETALDHx',
'BETALDHy',
'BSORx',
'BSORy',
'BTNt2ipp',
'BTNtex',
'BUTCT',
'BUTSO3abcpp',
'BUTSO3tex',
'BUTt2rpp',
'BUTtex',
'BWCOGDS1',
'BWCOGDS2',
'BWCOS',
'CAT',
'CBIAT',
'CBItonex',
'CBIuabcpp',
'CCGS',
'CD2tex',
'CDGR',
'CDGS',
'CFAS160E',
'CFAS160G',
'CFAS180E',
'CFAS180G',
'CHLabcpp',
'CHLt2pp',
'CHLtex',
'CHOLD',
'CHTBSptspp',
'CHTBStex',
'CINNDO',
'CMPtex',
'CPH4S',
'CRNBTCT',
'CRNCAL2',
'CRNCAR',
'CRNCBCT',
'CRNCDH',
'CRNDCAL2',
'CRNDtex',
'CRNt7pp',
'CRNtex',
'CSNt2pp',
'CSNtex',
'CTBTCAL2',
'CU1Opp',
'CU1abcpp',
'CUt3',
'CUtex',
'CYANST',
'CYANSTpp',
'CYANtex',
'CYNTAH',
'CYNTt2pp',
'CYNTtex',
'CYSDDS',
'CYSDabcpp',
'CYSDtex',
'CYSSADS',
'CYSTRS',
'DADNt2pp',
'DADNtex',
'DAMPtex',
'DAPAL',
'DAPabcpp',
'DC6PH',
'DCAtex',
'DCMPtex',
'DCYTt2pp',
'DCYTtex',
'DDCAtexi',
'DDGALK',
'DDGLCNt2rpp',
'DDGLCNtex',
'DDGLK',
'DDPGALA',
'DGMPtex',
'DGSNt2pp',
'DGSNtex',
'DHACOAH',
'DHCIND',
'DHCINDO',
'DHMPTR',
'DHNPTE',
'DHPPD',
'DHPTDNR',
'DHPTDNRN',
'DHPTPE',
'DIMPtex',
'DINSt2pp',
'DINStex',
'DKGLCNR1',
'DKGLCNR2x',
'DKGLCNR2y',
'DMSOR1',
'DMSOR1pp',
'DMSOR2',
'DMSOR2pp',
'DMSOtex',
'DMSOtpp',
'DMStex',
'DOGULNR',
'DOPAtex',
'DSBAO1',
'DSBAO2',
'DSBCGT',
'DSBDR',
'DSBGGT',
'DSERt2pp',
'DSERtex',
'DTARTD',
'DTMPtex',
'DUMPtex',
'DURADx',
'DURIt2pp',
'DURItex',
'DXYLK',
'ETHSO3abcpp',
'ETHSO3tex',
'F6Pt6_2pp',
'F6Ptex',
'FACOAL100t2pp',
'FACOAL80t2pp',
'FALDH2',
'FALDtex',
'FALDtpp',
'FALGTHLs',
'FCI',
'FCLK',
'FCLPA',
'FDMO',
'FDMO2',
'FDMO3',
'FDMO4',
'FDMO6',
'FE3DCITabcpp',
'FE3DCITtonex',
'FE3DHBZR',
'FE3DHBZSabcpp',
'FE3DHBZStonex',
'FESD2s',
'FFSD',
'FHL',
'FMETTRS',
'FORCT',
'FRUK',
'FRULYSDG',
'FRULYSE',
'FRULYSK',
'FRULYSt2pp',
'FRULYStex',
'FRUURt2rpp',
'FRUURtex',
'FRUpts2pp',
'FRUptspp',
'FRUtex',
'FUCtex',
'FUCtpp',
'FUMt2_2pp',
'FUMt2_3pp',
'FUMtex',
'G1PPpp',
'G1Ptex',
'G2PP',
'G2PPpp',
'G3PCabcpp',
'G3PCtex',
'G3PIabcpp',
'G3PItex',
'G3PSabcpp',
'G3PStex',
'G6Pt6_2pp',
'G6Ptex',
'GAL1PPpp',
'GAL1Ptex',
'GALBDtex',
'GALCTD',
'GALCTLO',
'GALCTND',
'GALCTNLt2pp',
'GALCTNLtex',
'GALCTNt2pp',
'GALCTNtex',
'GALCTt2rpp',
'GALCTtex',
'GALKr',
'GALM2pp',
'GALS3',
'GALTptspp',
'GALTtex',
'GALURt2rpp',
'GALURtex',
'GALabcpp',
'GALt2pp',
'GALtex',
'GAM6Pt6_2pp',
'GAMAN6Ptex',
'GAMptspp',
'GAMtex',
'GBBTNtex',
'GDMANE',
'GDPtex',
'GHBDHx',
'GLCATr',
'GLCRAL',
'GLCRD',
'GLCRt2rpp',
'GLCRtex',
'GLCUR1Ptex',
'GLCURt2rpp',
'GLCURtex',
'GLNTRS',
'GLNabcpp',
'GLNtex',
'GLTPD',
'GLUNpp',
'GLYBabcpp',
'GLYBt2pp',
'GLYBtex',
'GLYC2Pabcpp',
'GLYC2Ptex',
'GLYTRS',
'GMAND',
'GMPtex',
'GOFUCR',
'GP4GH',
'GPDDA1',
'GPDDA1pp',
'GPDDA3',
'GPDDA3pp',
'GPDDA5',
'GPDDA5pp',
'GSNt2pp',
'GSNtex',
'GTHOXtex',
'GTPtex',
'GUI1',
'GUI2',
'GUR1PPpp',
'H2O2tex',
'H2tex',
'H2tpp',
'HADPCOADH3',
'HCINNMt2rpp',
'HCINNMtex',
'HCYSMT2',
'HDCAtexi',
'HDCEAtexi',
'HETZK',
'HG2abcpp',
'HG2t3pp',
'HG2tex',
'HISTRS',
'HKNDDH',
'HKNTDH',
'HMPK1',
'HOPNTAL',
'HPPPNDO',
'HPPPNt2rpp',
'HPPPNtex',
'HYD1pp',
'HYD2pp',
'HYD3pp',
'ILETRS',
'IMPtex',
'INOSTt4pp',
'INSTtex',
'ISETACabcpp',
'ISETACtex',
'KG6PDC',
'LACZ',
'LACZpp',
'LALADGLUtex',
'LALADGLUtpp',
'LALALGLUtex',
'LALALGLUtpp',
'LCTStex',
'LEUTRS',
'LIPAHT2ex',
'LIPAHTex',
'LIPAMPL',
'LIPATPT',
'LIPOt2pp',
'LIPOtex',
'LPLIPAL2ATE120',
'LPLIPAL2ATE140',
'LPLIPAL2ATE141',
'LPLIPAL2ATE160',
'LPLIPAL2ATE161',
'LPLIPAL2ATE180',
'LPLIPAL2ATE181',
'LPLIPAL2ATG120',
'LPLIPAL2ATG140',
'LPLIPAL2ATG141',
'LPLIPAL2ATG160',
'LPLIPAL2ATG161',
'LPLIPAL2ATG180',
'LPLIPAL2ATG181',
'LYSTRS',
'LYXI',
'LYXt2pp',
'LYXtex',
'M1PD',
'MALDDH',
'MALDt2_2pp',
'MALDtex',
'MALTATr',
'MALTHXabcpp',
'MALTHXtexi',
'MALTPTabcpp',
'MALTPTtexi',
'MALTTRabcpp',
'MALTTRtexi',
'MALTTTRabcpp',
'MALTTTRtexi',
'MALTabcpp',
'MALTptspp',
'MALTtexi',
'MAN6PI',
'MAN6Pt6_2pp',
'MAN6Ptex',
'MANAO',
'MANGLYCptspp',
'MANGLYCtex',
'MANPGH',
'MANptspp',
'MANtex',
'MCPST',
'MELIBtex',
'METDabcpp',
'METDtex',
'METSOX1abcpp',
'METSOX1tex',
'METSOX2abcpp',
'METSOX2tex',
'METTRS',
'METabcpp',
'METtex',
'MI1PP',
'MINOHPtexi',
'MLTG1',
'MLTG2',
'MMETt2pp',
'MMETtex',
'MNLptspp',
'MNLtex',
'MNNH',
'MSO3abcpp',
'MSO3tex',
'MTRPOX',
'N2Otex',
'N2Otpp',
'NACtex',
'NACtpp',
'NAtex',
'NHFRBO',
'NMNPtpp',
'NMNt7pp',
'NMNtex',
'NNDMBRT',
'NO2t2rpp',
'NO2tex',
'NO3R1bpp',
'NO3R1pp',
'NO3R2bpp',
'NO3R2pp',
'NO3t7pp',
'NO3tex',
'NODOx',
'NODOy',
'NOtex',
'NOtpp',
'NTD10pp',
'NTD11pp',
'NTD12pp',
'NTD1pp',
'NTD2pp',
'NTD3pp',
'NTD4pp',
'NTD5pp',
'NTD6pp',
'NTD7pp',
'NTD8pp',
'NTD9pp',
'NTP11',
'NTP12',
'NTP3pp',
'NTRIR2x',
'NTRIR3pp',
'NTRIR4pp',
'O16A4COLIPAabctex',
'O16A4Lpp',
'O16AP1pp',
'O16AP2pp',
'O16AP3pp',
'O16AT',
'O16AUNDtpp',
'O16GALFT',
'O16GLCT1',
'O16GLCT2',
'O2Stex',
'OCDCAtexi',
'OCDCEAtexi',
'OCTAtex',
'OP4ENH',
'OROTt2_2pp',
'OROTtex',
'OXCDC',
'OXCOAHDH',
'OXDHCOAT',
'PACALDt2rpp',
'PACALDtex',
'PACCOAE',
'PACCOAL',
'PEAMNOpp',
'PEAMNtex',
'PFK_2',
'PGLYCP',
'PHETRS',
'PHYTSpp',
'PNTOt4pp',
'PNTOtex',
'PPALtex',
'PPALtpp',
'PPAt4pp',
'PPAtex',
'PPPNDO',
'PPPNt2rpp',
'PPPNtex',
'PPTHpp',
'PPTtex',
'PROGLYabcpp',
'PROGLYtex',
'PROTRS',
'PSCLYSt2pp',
'PSCLYStex',
'PSERtex',
'PSP_Lpp',
'PTHRpp',
'PYDAMtex',
'PYDAMtpp',
'PYDXNtex',
'PYDXNtpp',
'PYDXtex',
'PYDXtpp',
'QMO2',
'QMO3',
'R5PPpp',
'R5Ptex',
'RBK_L1',
'RBP4E',
'REPHACCOAI',
'RIBabcpp',
'RIBtex',
'RMI',
'RMK',
'RMNtex',
'RMNtpp',
'RMPA',
'RZ5PP',
'SARCOX',
'SBTPD',
'SBTptspp',
'SBTtex',
'SELCYSS',
'SELGTHR',
'SELGTHR2',
'SELGTHR3',
'SELNPS',
'SELR',
'SELtex',
'SELtpp',
'SERTRS',
'SERTRS2',
'SFGTHi',
'SKMt2pp',
'SKMtex',
'SLNTtex',
'SLNTtpp',
'SO2tex',
'SO2tpp',
'SO3tex',
'SPMDAT1',
'SPMDAT2',
'SPODM',
'SPODMpp',
'SUCASPtpp',
'SUCFUMtpp',
'SUCMALtpp',
'SUCRtex',
'SUCTARTtpp',
'SUCptspp',
'SULFACabcpp',
'SULFACtex',
'TAGURr',
'TARTD',
'TARTRDtex',
'TARTRt7pp',
'TARTRtex',
'TARTt2_3pp',
'TAUDO',
'TAURabcpp',
'TAURtex',
'TCYNTtex',
'TDSR1',
'TDSR2',
'TGBPA',
'THMabcpp',
'THMtex',
'THRPtex',
'THRTRS',
'TMAOR1',
'TMAOR1pp',
'TMAOR2',
'TMAOR2pp',
'TMAOtex',
'TMAtex',
'TMK',
'TPRDCOAS',
'TREHpp',
'TREptspp',
'TREtex',
'TRPTRS',
'TSULabcpp',
'TSULtex',
'TTDCAtexi',
'TTDCEAtexi',
'TUNGSabcpp',
'TUNGStex',
'TYMtex',
'TYROXDApp',
'TYRPpp',
'TYRPtex',
'TYRTRS',
'ThDPAT',
'UACGALPpp',
'UACGAMPpp',
'UACGAMtex',
'UDPACGALtex',
'UDPG4E',
'UDPGALM',
'UDPGALPpp',
'UDPGALtex',
'UDPGLCURtex',
'UDPGPpp',
'UDPGtex',
'UGLCURPpp',
'UGLT',
'UMPtex',
'VALTRS',
'WCOS',
'X5PL3E',
'XMPtex',
'XYLI1',
'XYLK',
'XYLK2',
'XYLUt2pp',
'XYLUtex',
'XYLabcpp',
'XYLt2pp',
'XYLtex']