Gillespie Algorithm¶
using CairoMakie
using DataInterpolations: LinearInterpolation
using StatsBase: Weights, sample
using Statistics: mean
using Random
Random.seed!(2024)Random.TaskLocalRNG()Do-it-yourself¶
Stochastic chemical reaction: Gillespie Algorithm (direct and first reaction method) Adapted from: Chemical and Biomedical Engineering Calculations Using Python Ch.4-3
function ssa_alg(model, u0::AbstractVector, tend, p, stoich; tstart=zero(tend), method=:direct)
t = tstart ## Current time
ts = [t] ## Time points
u = copy(u0) ## Current state
us = copy(u') ## States over time
while t < tend
a = model(u, p, t) ## propensities
if method == :direct
dt = randexp() / sum(a) ## Time step for the direct method
du = sample(stoich, Weights(a)) ## Choose the stoichiometry for the next reaction
elseif method == :first
dts = randexp(length(a)) ./ a ## time scales of all reactions
i = argmin(dts) ## Choose the most recent reaction to occur
dt = dts[i]
du = stoich[i]
else
error("Method should be either :direct or :first")
end
u .+= du ## Update time
t += dt ## Update time
us = [us; u'] ## Append state
push!(ts, t) ## Append time point
end
return (t=ts, u=us)
endssa_alg (generic function with 1 method)Propensity model for this example reaction. Reaction of A <-> B with rate constants k1 & k2
model(u, p, t) = [p.k1 * u[1], p.k2 * u[2]]model (generic function with 1 method)parameters = (k1=1.0, k2=0.5)(k1 = 1.0, k2 = 0.5)Stoichiometry for each reaction
stoich = [[-1, 1], [1, -1]]2-element Vector{Vector{Int64}}:
[-1, 1]
[1, -1]Initial conditions (Usually discrete values)
u0 = [200, 0]2-element Vector{Int64}:
200
0Simulation time
tend = 10.010.0Solve the problem using both direct and first reaction method
@time soldirect = ssa_alg(model, u0, tend, parameters, stoich; method=:direct)
@time solfirst = ssa_alg(model, u0, tend, parameters, stoich; method=:first) 0.186343 seconds (688.01 k allocations: 61.390 MiB, 7.54% gc time, 98.37% compilation time)
0.012432 seconds (9.95 k allocations: 14.957 MiB, 88.65% gc time)
(t = [0.0, 0.006874672879606227, 0.0070050915014185, 0.01329115182676454, 0.015335530116774018, 0.01942319769041779, 0.021346508038833215, 0.02345030020299408, 0.025497753122539372, 0.02734483697039726 … 9.937773197389458, 9.938360255993572, 9.964450740930666, 9.96564917153764, 9.975440668199402, 9.978594329657025, 9.982318425245289, 9.987213207313044, 9.995833233887383, 10.024199143896238], u = [200 0; 199 1; … ; 75 125; 74 126])Plot the solution from the direct method
fig, ax, sp = series(soldirect.t, soldirect.u', labels=["A", "B"]; axis=(; xlabel="time", ylabel="# of molecules", title="SSA (direct method)"))
axislegend(ax)
fig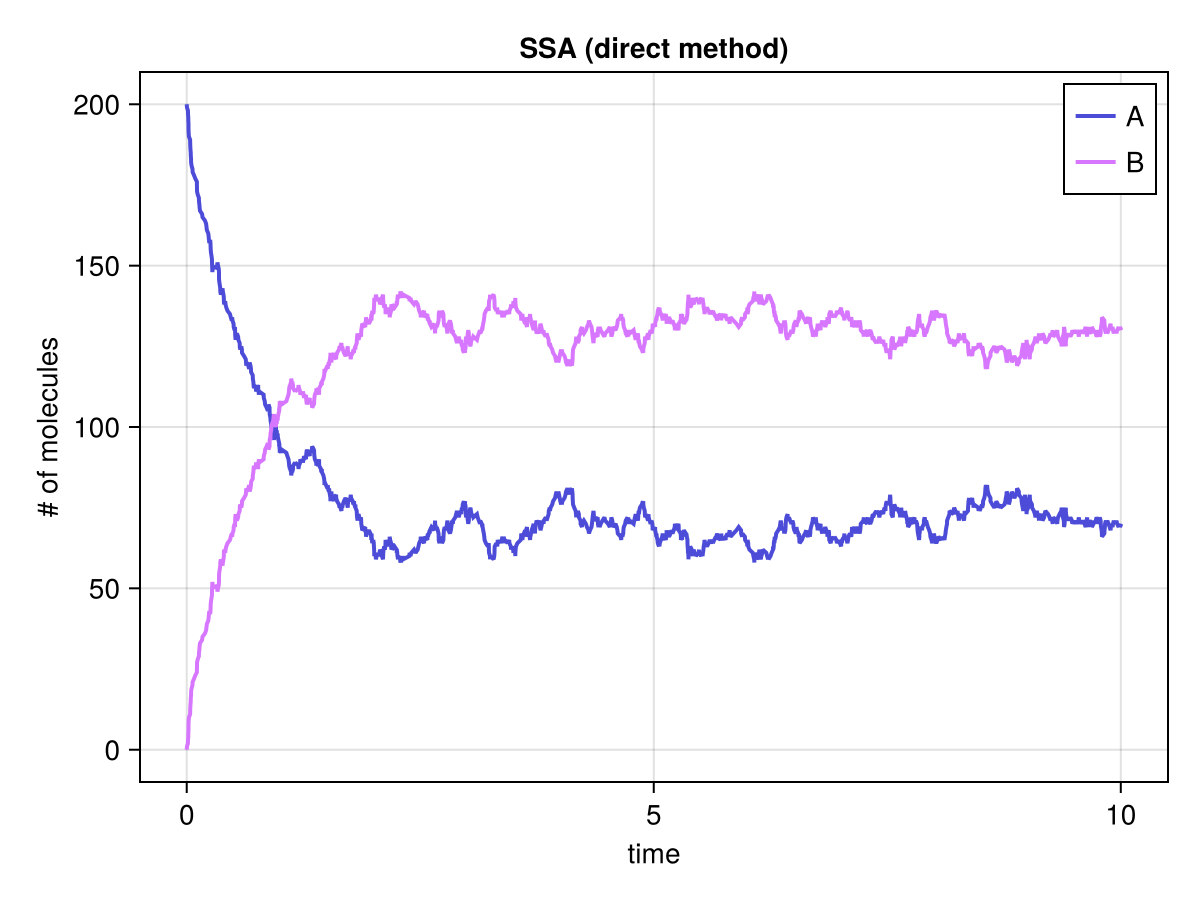
Plot the solution by the first reaction method
fig, ax, sp = series(solfirst.t, solfirst.u', labels=["A", "B"]; axis=(; xlabel="time", ylabel="# of molecules", title="SSA (first reaction method)"))
axislegend(ax)
fig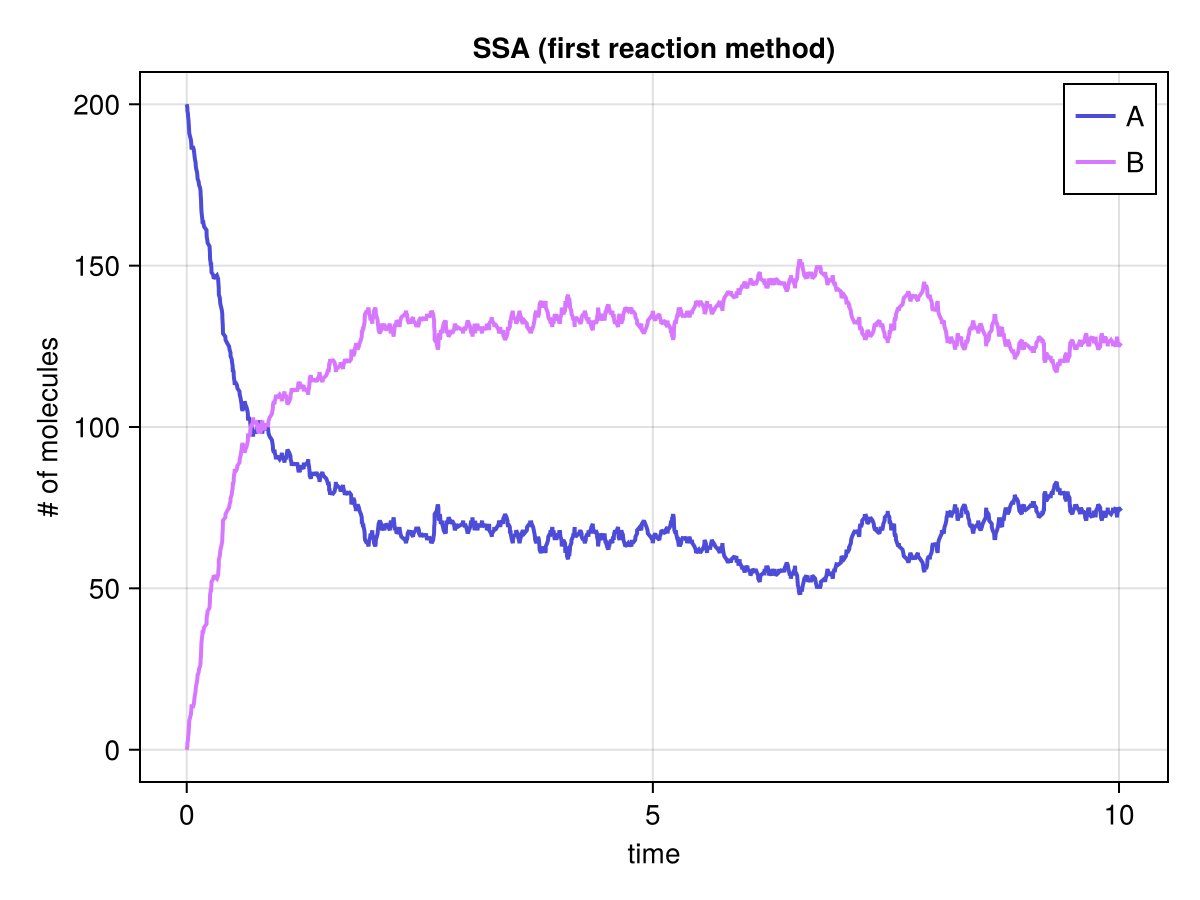
Running 50 simulations
@time sols = map(1:50) do _
ssa_alg(model, u0, tend, parameters, stoich; method=:direct)
end; 0.133689 seconds (635.51 k allocations: 760.284 MiB, 24.36% gc time, 39.54% compilation time)
Average values and interpolation
ts = range(0, tend, 101)
a_interp = [LinearInterpolation(sol.u[:, 1], sol.t; cache_parameters=true) for sol in sols]
b_interp = [LinearInterpolation(sol.u[:, 2], sol.t; cache_parameters=true) for sol in sols]
a_avg(t) = mean(i-> i(t), a_interp)
b_avg(t) = mean(i-> i(t), b_interp)b_avg (generic function with 1 method)Plot the solution
fig = Figure()
ax = Axis(fig[1, 1],
xlabel = "Time",
ylabel = "# of molecules",
title = "SSA (first method) ensemble"
)
for sol in sols
lines!(ax, sol.t, sol.u[:, 1], color = (:blue, 0.05))
lines!(ax, sol.t, sol.u[:, 2], color = (:red, 0.05))
end
lines!(ax, ts, a_avg.(ts), color = :black, linewidth = 3, linestyle = :solid, label = "Average [A]")
lines!(ax, ts, b_avg.(ts), color = :black, linewidth = 3, linestyle = :dash, label = "Average [B]")
axislegend(ax, position = :rt)
fig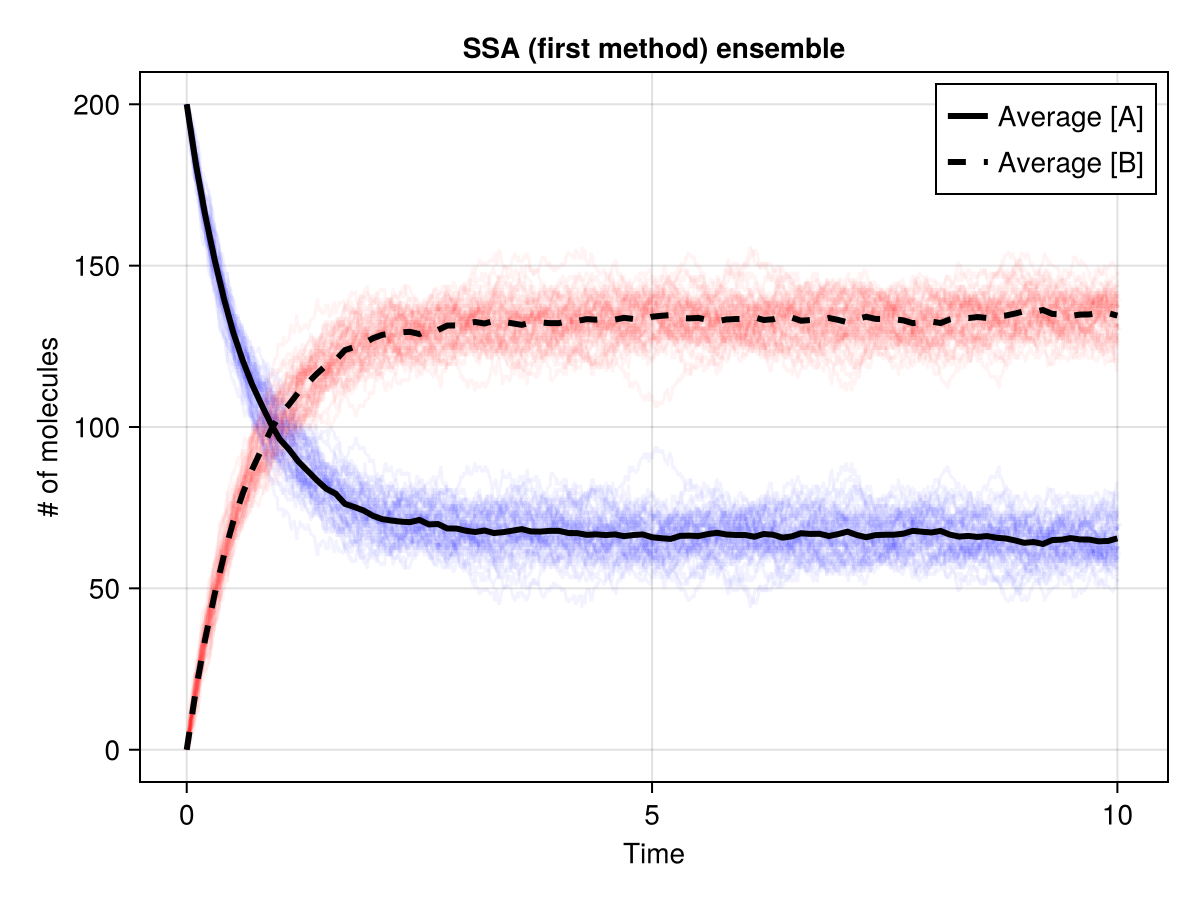
This notebook was generated using Literate.jl.