stochastic implementation (Gillespie SSA) of a collection of spontaneously decaying molecules.
using CairoMakie
using JumpProcesses
using ComponentArrays: ComponentArrayDefine propensity functions and effects
v1_738(u, p, t) = p.d * u[1]
function affectv1_738!(integrator)
integrator.u[1] -= 1 ## A -> 01
nothing
endaffectv1_738! (generic function with 1 method)ps738 = ComponentArray(d=1)
tend = 5.0
jump1 = ConstantRateJump(v1_738, affectv1_738!)
prob1000 = DiscreteProblem(ComponentArray(A=1000), (0.0, tend), ps738)
jump_prob1000 = JumpProblem(prob1000, Direct(), jump1)
@time sol1000 = solve(jump_prob1000, SSAStepper())
prob100 = DiscreteProblem(ComponentArray(A=100), (0.0, tend), ps738)
jump_prob100 = JumpProblem(prob100, Direct(), jump1)
@time sol100 = solve(jump_prob100, SSAStepper())
prob10 = DiscreteProblem(ComponentArray(A=10), (0.0, tend), ps738)
jump_prob10 = JumpProblem(prob10, Direct(), jump1)
@time sol10 = solve(jump_prob10, SSAStepper()) 0.130343 seconds (538.19 k allocations: 36.595 MiB, 99.95% compilation time)
0.000019 seconds (114 allocations: 14.656 KiB)
0.000013 seconds (22 allocations: 3.156 KiB)
retcode: Success
Interpolation: Piecewise constant interpolation
t: 12-element Vector{Float64}:
0.0
0.04356189666580788
0.2435413916819835
0.24942631732244896
0.38356786238367435
0.8215887908345435
0.9754552997134038
1.03030370289381
1.1444142269887034
1.4072903307503797
1.8876750555471262
5.0
u: 12-element Vector{ComponentArrays.ComponentVector{Int64, Vector{Int64}, Tuple{ComponentArrays.Axis{(A = 1,)}}}}:
ComponentVector{Int64}(A = 10)
ComponentVector{Int64}(A = 9)
ComponentVector{Int64}(A = 8)
ComponentVector{Int64}(A = 7)
ComponentVector{Int64}(A = 6)
ComponentVector{Int64}(A = 5)
ComponentVector{Int64}(A = 4)
ComponentVector{Int64}(A = 3)
ComponentVector{Int64}(A = 2)
ComponentVector{Int64}(A = 1)
ComponentVector{Int64}(A = 0)
ComponentVector{Int64}(A = 0)fig = Figure()
ax = Axis(fig[1, 1],
xlabel="Time",
ylabel="# of molecules",
title="A"
)
lines!(ax, sol1000.t, reduce(vcat, sol1000.u))
lines!(ax, 0..tend, t->1000 * exp(-ps738.d * t), linestyle=:dash)
limits!(ax, 0, tend, 0, 1000)
ax = Axis(fig[1, 2],
xlabel="Time",
ylabel="# of molecules",
title="B"
)
lines!(ax, sol100.t, reduce(vcat, sol100.u))
lines!(ax, 0..tend, t->100 * exp(-ps738.d * t), linestyle=:dash)
limits!(ax, 0, tend, 0, 100)
ax = Axis(fig[1, 3],
xlabel="Time",
ylabel="# of molecules",
title="C"
)
lines!(ax, sol10.t, reduce(vcat, sol10.u))
lines!(ax, 0..tend, t->10 * exp(-ps738.d * t), linestyle=:dash)
limits!(ax, 0, tend, 0, 10)
fig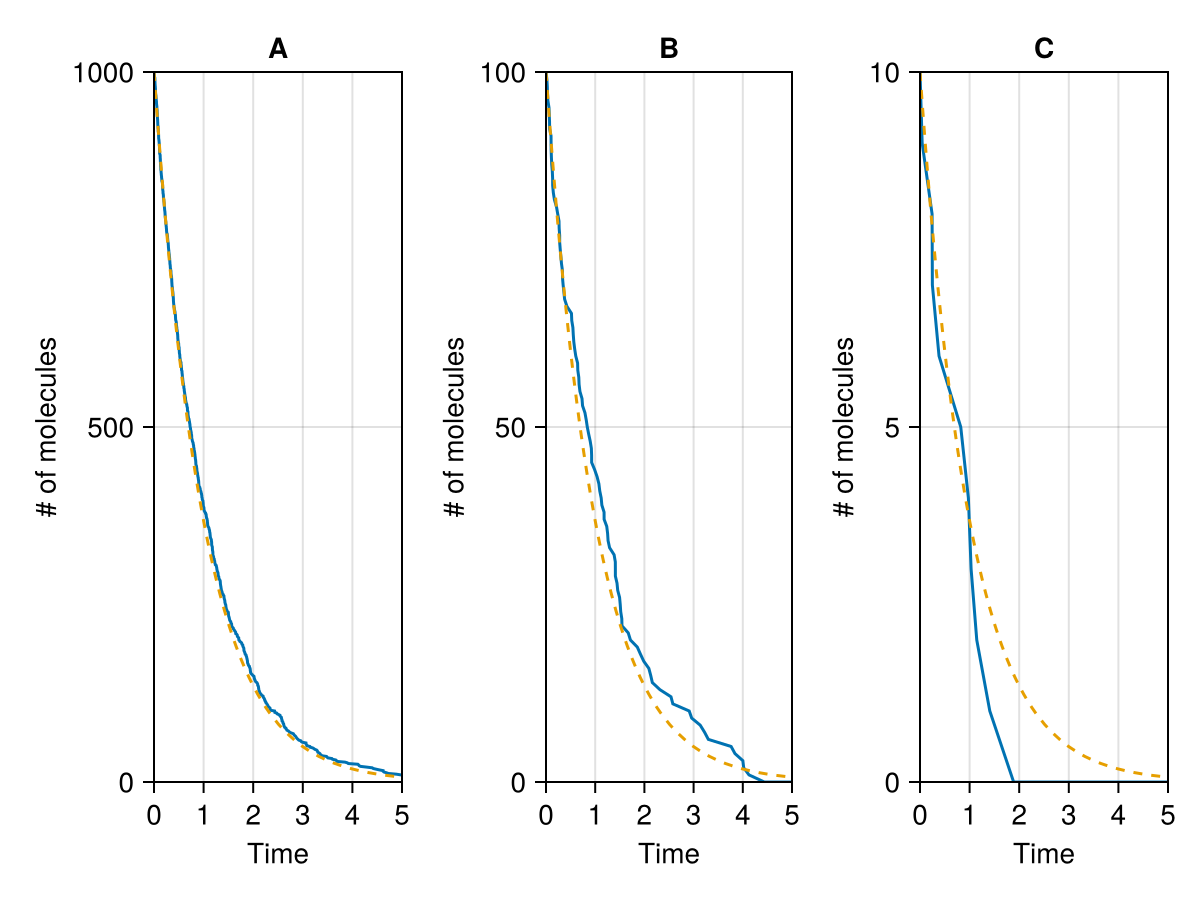
This notebook was generated using Literate.jl.